Melenis repens associated virus
Basic Information
| Genus | Maldovirus |
|---|---|
| Isolate | Reunion |
| Host | |
| Vector | |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
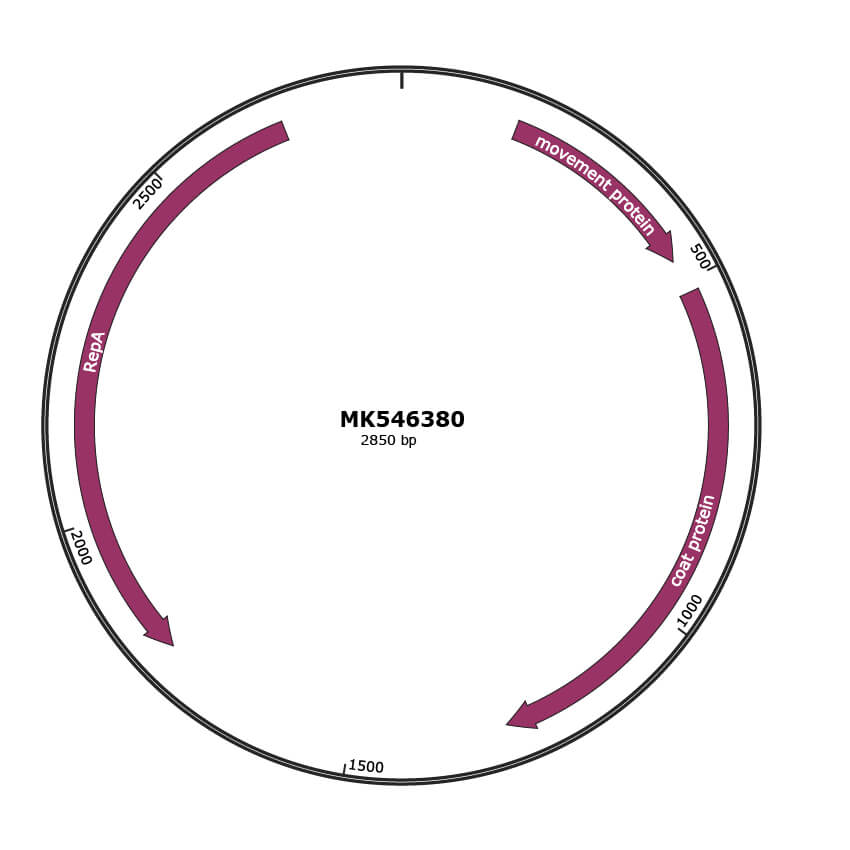
JBrowse
Genome
MK546380
Gene Information
| NCBI Accession | QDO73336.1 |
|---|---|
| Location | 168-467 |
| Protein Name | movement protein |
| Coding Region | ATGGAGAAGGGTCCACTTCCGCAGGAATTTACAGTGGGGTCTAATCTCCCTCCCGCCGGGCCCTCAGGCGTCGGTAACGACGCTGCGTGGAGGTTCTTGGTTTTACTCTTCGCGTGTACTGCGGTTGCTGGTGTCTCGGTCTTCGCCTGCTGGAGGCTCTGCGGTAAGGATCTGCTCCTTACTCTGAAGGCAAAGCGTAGCAGGACCATCACGGAGCTAGGGTTCGGCCAGACTCCCGAAGGGAGGTACGCTGGAAGGGGTGCTCAACAGGTGGGCCAGGTCGGCGGGATCCCGGGGTAG |
| Protein Sequence | MEKGPLPQEFTVGSNLPPAGPSGVGNDAAWRFLVLLFACTAVAGVSVFACWRLCGKDLLLTLKAKRSRTITELGFGQTPEGRYAGRGAQQVGQVGGIPG |
| NCBI Accession | QDO73337.1 |
|---|---|
| Location | 517-1272 |
| Protein Name | coat protein |
| Coding Region | ATGCCGGCGGCGAGGGGGAGCACCAGTAGAAAGAGGACTAGGGATGACTGGGCCTGGGACTCTCGCAAGAGGCGTCGTGTTCGGCCCACTACGTCTGCTATTCCGCGTGGGCCTCGTGGCCGTAGGCCCAGTTTGCAGGTGGATCAGGACGTATATGCAGGGCCCACTCTCGTTGAAGTTCCCGCCGTTGGCGGGATAGTTGGTATTATGACCAACTTCATGCGGGGGAGTGACGAGAGCGCGCGTCACACTGCAGAGACTATCCTGTACAAGATAGGGATCGATGTGTATGTGACTGTCTCGGAGGCCCTTACAGTATATTCAGGTAAGGGGGTCAATGTCGCTTGGCTGGTATACGATGCGCAGCCGACTGGCACGCGTCCTACGTGTGCCGATATCTTCCCGCTGGCGAGTCCGAATTTCAATACTCAGCCGTGTGTATGGAAAGTGGGAAGGGAGGTCTGCCATCGTTTTGTGGTTAAGAGGAGGTGGCTTTTCACTCTGGAGACCAATGGAGTTCGTCCTGGCACTTCGTTCTCGGGGGCGAACGGCGTTCCCCCATGTAATAGATCCCTGTACTTCCACAAGTTCTGCAAGCGATTGGGCGTCAGGACGGAGTGGAAGAACACCCCCGGAGGGACAGATGGGGACATCAAGACGGGTGCCTTGTATCTAGTATTCGCTGCGGGCAATAAGATGTCCTTCAGCGTTAGCGGGGTATGCAGGAAGTACTTCAAGAGTGTCGGGAACCAGTAG |
| Protein Sequence | MPAARGSTSRKRTRDDWAWDSRKRRRVRPTTSAIPRGPRGRRPSLQVDQDVYAGPTLVEVPAVGGIVGIMTNFMRGSDESARHTAETILYKIGIDVYVTVSEALTVYSGKGVNVAWLVYDAQPTGTRPTCADIFPLASPNFNTQPCVWKVGREVCHRFVVKRRWLFTLETNGVRPGTSFSGANGVPPCNRSLYFHKFCKRLGVRTEWKNTPGGTDGDIKTGALYLVFAAGNKMSFSVSGVCRKYFKSVGNQ |
| NCBI Accession | QDO73338.1 |
|---|---|
| Location | 1790-2680 |
| Protein Name | RepA |
| Coding Region | ATGAACACTGAGCCAGGAGAGTCTAGCCGATTCTACTTCCAAAATTCAAATGTCTTCCTCACATACTCCAGATGCCCACTGGAGCCTGAGGTTGTGGGAAGCAATCTCAAAAACCTCCTTTCAGCTTACGCTGTTAGTTTCGTCGCTGTTAATCGCGAGCTCCATCAAGATGGGACTCCACACATCCACGCCCTGGCTCAGACTAATCAACGTATCTATACATCTGATACAGGCTTCTTTGATATTCTGGGTTTTCACCCTAACATACAAGGTACTCGAAACTGCAAAGCTGTGCTGGCTTATATATCTAAATACTCTTTGGGAGAATTCCGTTGGGGTACCTTCCGGGCCCGCGGAGCACGCGCACCTACTGCACGTGCAGAGGGAGCGGTTCAAGCTGATGCAGGATCTTCAAACAATTCTCGACCTGCCGCCGATAGTGAACCTGGATCAGGAGGATCCGGGCCTATACGACACAGACGAGCTCCCCGACAGCCCTTGAAGGACGAGGTGATGGCGTCCATCCTCCGGCGGTGCTCCTCCAGGCAGGAGTACCTCAATGAGGTCAAGAAGTGCTTCCCGTACGACTTCTGCGTCCGGCTTCAAGCCTGGGAGTACGCTGCTCAGAAGCTCTACCCCGATCCACCTTCGGCATACGAACCTCCATTTCCGGACTCCTTGTTTCGCTGCCACGAAACACTGTCCGATTGGGTCCGTGATAATGTCTACCAGGTCACTCCGGAGGTATACCAACTTCTTCACCCTCACGCGGACGCTGAAGCTGAACTTAGATGGCTCCATGACACATGCTTTGAAAGCAGGTGCGACCGAGCTCCCTCTACATCTGCGGCCCAACAAGGACAGGGAAGACAACATGGGCCCGAAGCTTAG |
| Protein Sequence | MNTEPGESSRFYFQNSNVFLTYSRCPLEPEVVGSNLKNLLSAYAVSFVAVNRELHQDGTPHIHALAQTNQRIYTSDTGFFDILGFHPNIQGTRNCKAVLAYISKYSLGEFRWGTFRARGARAPTARAEGAVQADAGSSNNSRPAADSEPGSGGSGPIRHRRAPRQPLKDEVMASILRRCSSRQEYLNEVKKCFPYDFCVRLQAWEYAAQKLYPDPPSAYEPPFPDSLFRCHETLSDWVRDNVYQVTPEVYQLLHPHADAEAELRWLHDTCFESRCDRAPSTSAAQQGQGRQHGPEA |
References More References in PubMed
| 1 |
Dirofilaria spp. infection in cats from the Mediterranean basin: diagnosis and epidemiology. Carbonara M, et al. Int J Parasitol. 2025 May;55(6):317-325. doi: 10.1016/j.ijpara.2025.01.011. Epub 2025 Feb 3. PMID: 39909190 |
|---|---|
| 2 |
Retention and Transmission of Grapevine Leafroll-Associated Virus 3 by Pseudococcus calceolariae. McGreal B, et al. Front Microbiol. 2021 May 12;12:663948. doi: 10.3389/fmicb.2021.663948. eCollection 2021. PMID: 34054767 |
| 3 |
Abdalla OA, et al. Plant Dis. 2012 Nov;96(11):1705. doi: 10.1094/PDIS-07-12-0630-PDN. PMID: 30727477 |
| 4 |
Genomic characterization of a novel potyvirus infecting Barleria repens in South Africa. Read DA, et al. Arch Virol. 2022 Dec 28;168(1):10. doi: 10.1007/s00705-022-05662-w. PMID: 36576587 |
| 5 |
Association of phytoplasmas and viruses with malformed clovers. Fránová J, et al. Folia Microbiol (Praha). 2004;49(5):617-24. doi: 10.1007/BF02931544. PMID: 15702556 |
| 6 |
Hampton RO, et al. J Econ Entomol. 2005 Dec;98(6):1816-23. doi: 10.1603/0022-0493-98.6.1816. PMID: 16539099 |
| 7 |
Maclot FJ, et al. Virus Res. 2021 Jun;298:198397. doi: 10.1016/j.virusres.2021.198397. Epub 2021 Mar 18. PMID: 33744338 |
| 8 |
A cross section through mosquitoes of Bosnia and Herzegovina: Barcodes, blood meals and pathogens. Hoxha I, et al. One Health. 2025 Oct 15;21:101246. doi: 10.1016/j.onehlt.2025.101246. eCollection 2025 Dec. PMID: 41159105 |
| 9 |
Biddle JM, et al. Ecol Appl. 2012 Jan;22(1):35-52. doi: 10.1890/11-0341.1. PMID: 22471074 |
| 10 |
van Mölken T, et al. Ann Bot. 2011 Jun;107(8):1391-7. doi: 10.1093/aob/mcr078. Epub 2011 Apr 22. PMID: 21515605 |