Chilli leaf curl virus
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_000841265.1 |
| Isolate | Pakistan: Multan, Punjab province |
| Release date | 2015/2/12 |
| Submitter | Shih,S.L., Tsai,W.S., Green,S.K., Khalid,S., Ahmad,I., Rezaian,M.A., Smith,J. |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
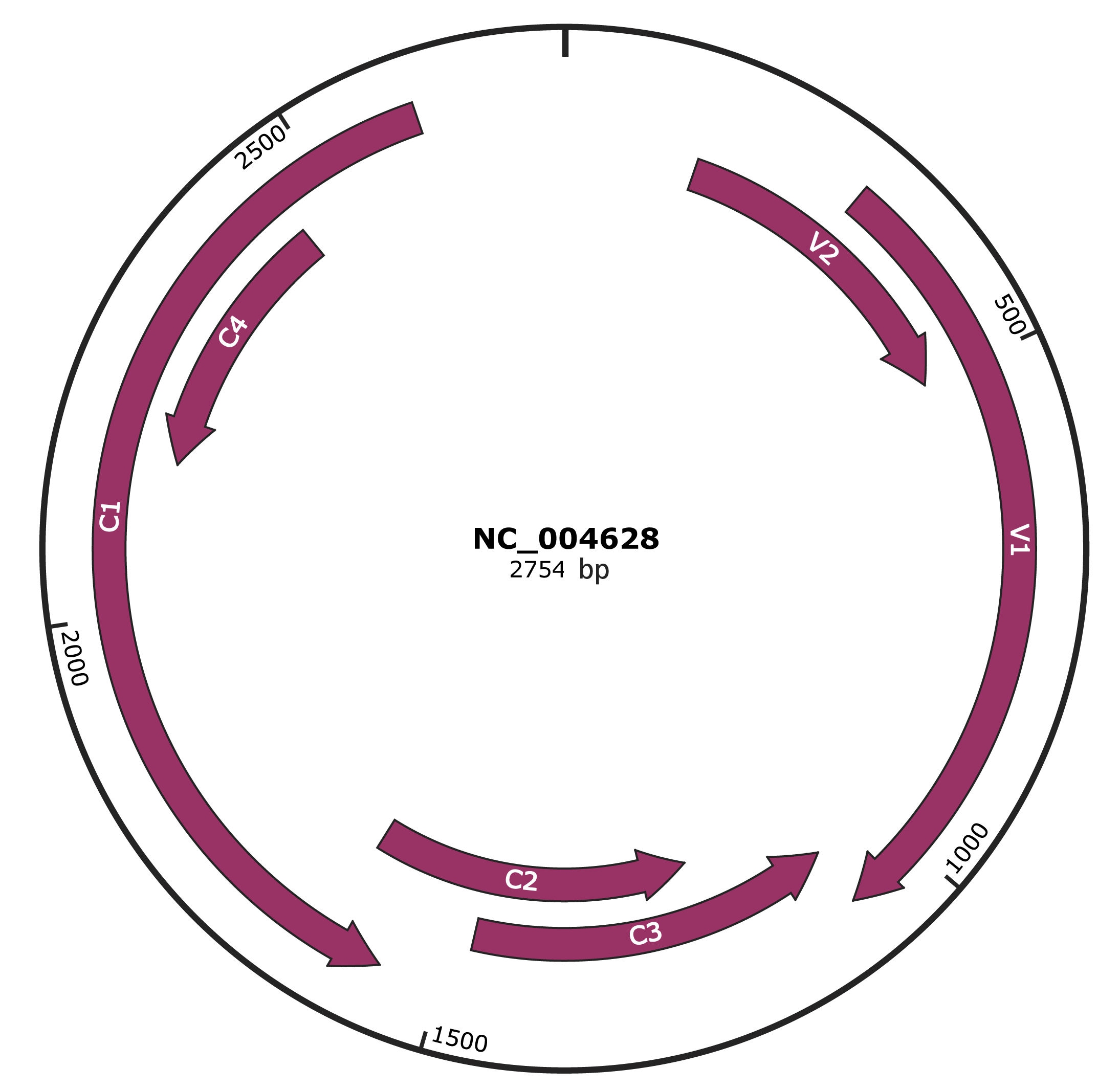
JBrowse
Genome
NC_004628
Gene Information
| NCBI Accession | NP_803553.1 |
|---|---|
| Location | 146-502 |
| Gene Name | V2 |
| Protein Name | pre-coat protein |
| Coding Region | ATGTGGGATCCATTAGTAAACGAGTTTCCTGAAACCGTTCACGGTTTTAGGTGTATGTTAGCAGTTAAACATCTGCAGCTACTAGAAAATACATATTCTCCAGACACTCTGGGGTACGATTTAATTCGGGATTTGATCTCCGTTATTAGGGCTAAGAATTATGTCCAAGCGACCAGCAGATATAATCATTTCCACGCCCGCCTCGAAGGTACGCCGCCGTCTCAACTTCGACAGCCCTTATGCCAGCCGTGCTGCTGCCCCCATTGTCCGCGTCACAAAGGCCAGAGCATGGGCGAACAGGCCCATGAACAGGAAGCCCAGGATGTACAGGATGTACAGAAGCCCAGATGTTCCTAG |
| Protein Sequence | MWDPLVNEFPETVHGFRCMLAVKHLQLLENTYSPDTLGYDLIRDLISVIRAKNYVQATSRYNHFHARLEGTPPSQLRQPLCQPCCCPHCPRHKGQSMGEQAHEQEAQDVQDVQKPRCS |
| NCBI Accession | NP_803554.1 |
|---|---|
| Location | 306-1076 |
| Gene Name | V1 |
| Protein Name | coat protein |
| Coding Region | ATGTCCAAGCGACCAGCAGATATAATCATTTCCACGCCCGCCTCGAAGGTACGCCGCCGTCTCAACTTCGACAGCCCTTATGCCAGCCGTGCTGCTGCCCCCATTGTCCGCGTCACAAAGGCCAGAGCATGGGCGAACAGGCCCATGAACAGGAAGCCCAGGATGTACAGGATGTACAGAAGCCCAGATGTTCCTAGGGGGTGTGAAGGCCCATGTAAGGTCCAGTCTTTTGAGTCTAGACACGATGTAGTTCATATAGGGAAGGTGATGTGTATTAGTGATGTTACCCGTGGTACTGGGTTAACCCATAGAGTAGGTAAGCGTTTCTGTGTTAAGTCTGTGTATGTATTAGGTAAGATATGGATGGATGAGAATATCAAGACTAAGAACCACACGAATAGTGTGATGTTCTTCCTTGTCCGTGACCGTCGTCCTGTTGATAAACCACAAGATTTTGGAGAGGTGTTCAACATGTTTGACAACGAGCCTAGCACTGCCACTGTGAAGAATATGCATAGAGATCGTTATCAGGTTTTGAGAAAATGGCACGCAACGGTTACTGGTGGACAGTACGCTTCGAAGGAACAGGCATTAGTTAAGAAGTTCGTTAGGGTTAACAATTATGTTGTGTATAACCAGCAAGAAGCTGGAAAATATGAGAATCATACTGAGAATGCATTGATGTTGTACATGGCGTGTACTCACGCCTCTAATCCTGTGTATGCTACGTTACAGATACGGATCTATTTCTATGATTCAGTATCGAATTAA |
| Protein Sequence | MSKRPADIIISTPASKVRRRLNFDSPYASRAAAPIVRVTKARAWANRPMNRKPRMYRMYRSPDVPRGCEGPCKVQSFESRHDVVHIGKVMCISDVTRGTGLTHRVGKRFCVKSVYVLGKIWMDENIKTKNHTNSVMFFLVRDRRPVDKPQDFGEVFNMFDNEPSTATVKNMHRDRYQVLRKWHATVTGGQYASKEQALVKKFVRVNNYVVYNQQEAGKYENHTENALMLYMACTHASNPVYATLQIRIYFYDSVSN |
| NCBI Accession | NP_803555.1 |
|---|---|
| Location | 1073-1477 |
| Gene Name | C3 |
| Protein Name | C3 protein |
| Coding Region | ATGGATTCACGCACAGGGGAACCCATCACTGCAGCTCAGGCAGAGAATGGCGTGTTTATCTGGGAGATAACAAATCCCCTGTATTTCAAGATAACCGAGCACCACAACAGGCCATTCCTGAGGAGCCGAGACCTCATCACCGTGCAGATACAGTTCAATCACAACCTGAGGAAAGCGTTGGGGATACACAAATGTTTTCTAACCTTCCGAATCTGGACGACCTTACAGCCTCAGACTGGTCATTTCTTAAGGGTATTTAAGACCCAAGTGTTGCAGTTTTTAAATAATTTTGCTGTAATTAGTATAAACAATGTAATTAGAGCTGTCGACCATGTATTATGGAATGTATTACATAACATTGTATATGTACAACAATCATATTCAATAAAATTCAATCTTTATTAA |
| Protein Sequence | MDSRTGEPITAAQAENGVFIWEITNPLYFKITEHHNRPFLRSRDLITVQIQFNHNLRKALGIHKCFLTFRIWTTLQPQTGHFLRVFKTQVLQFLNNFAVISINNVIRAVDHVLWNVLHNIVYVQQSYSIKFNLY |
| NCBI Accession | NP_803556.1 |
|---|---|
| Location | 1218-1622 |
| Gene Name | C2 |
| Protein Name | C2 protein |
| Coding Region | ATGCGACCTTCGTCACCCTCACAGGCCCACTCTACTCAGGTTCCAATCAAAGTACAGCATAGGCTAGCGAAGAAGGGGACCAGGCGTCGTCGCGTTGACCTAGACTGCGGCTGTTCATATTTCATCGCATTACGCTGCCACGGCTATGGATTCACGCACAGGGGAACCCATCACTGCAGCTCAGGCAGAGAATGGCGTGTTTATCTGGGAGATAACAAATCCCCTGTATTTCAAGATAACCGAGCACCACAACAGGCCATTCCTGAGGAGCCGAGACCTCATCACCGTGCAGATACAGTTCAATCACAACCTGAGGAAAGCGTTGGGGATACACAAATGTTTTCTAACCTTCCGAATCTGGACGACCTTACAGCCTCAGACTGGTCATTTCTTAAGGGTATTTAA |
| Protein Sequence | MRPSSPSQAHSTQVPIKVQHRLAKKGTRRRRVDLDCGCSYFIALRCHGYGFTHRGTHHCSSGREWRVYLGDNKSPVFQDNRAPQQAIPEEPRPHHRADTVQSQPEESVGDTQMFSNLPNLDDLTASDWSFLKGI |
| NCBI Accession | NP_803557.1 |
|---|---|
| Location | 1561-2610 |
| Gene Name | C1 |
| Protein Name | replication-associated protein |
| Coding Region | ATGCCTAGGGCTGGCAGATTCAATATAAATGCTAAAAATTATTTCCTAACATATCCCAACTGCTCTCTTACTAAAGAAGAGGCACTTTCCCAATTACAGAACTTGGAAACCCCAGTTAATAAATTATTTATTAGGGTTTGTCGAGAATTGCACGAGAATGGGGAACCTCATCTACACGTGCTCATCCAATTCGAGGGAAAATACCAATGCACAAACAACAGATTCTTCGACCTTATTTCCCCAACCCGGTCAGCAAATTTCCATCCGAACATTCAGAGAGCTAAAAGCTCTTCAGATGTCAAAGCCTATGTGGAGAAAGACGGAGACTTCATTGACTTTGGAGTTTTCCAAATCGATGGCAGATCAGCTAGAGGAGGTTGCCAATCTGCCAACGACGCATATGCCGAGGCAATCAATTCAGGGTCCAAGTCTTCGGCCCTCAATATTTTAAGGGAGAAAGCTCCCAAAGATTATGTTTTACAATTTCATAATTTAAATGCTAATTTAGATAGGATTTTTACACCTCCCTTGGAGGTTTATGTTTCTCCTTTTTGTTCTTCTTCTTTTGATCAAGTTCCCGAGGAACTTGAGGAGTGGGCTGCCGAGAATGTTCTCGGTGCCGCTGCGCGGCCTTTAAGACCCATAAGTATTGTCATAGAGGGTGAGAGTAGGACGGGGAAGACGATATGGGCCAGGTCATTGGGTCCACACAACTATTTGTGTGGACATCTCGACCTTAGCCCAAAGGTCTACAGTAATGACGCCTGGTACAACGTCATTGATGACGTCGATCCCCACTACCTAAAGCACTTTAAGGAATTCATGGGGGCCCAAAGGGACTGGCAATCAAATACTAAATACGGGAAGCCAGTTCAAATTAAAGGTGGAATTCCCGCTATCTTCCTCTGCAATCCAGGACCCAATTCCAGCTATAAAGAGTTCTTAGACGAAGAGAAAAATAGCGCACTCAAAAATTGGGCTTTGAAGAATGCGACCTTCGTCACCCTCACAGGCCCACTCTACTCAGGTTCCAATCAAAGTACAGCATAG |
| Protein Sequence | MPRAGRFNINAKNYFLTYPNCSLTKEEALSQLQNLETPVNKLFIRVCRELHENGEPHLHVLIQFEGKYQCTNNRFFDLISPTRSANFHPNIQRAKSSSDVKAYVEKDGDFIDFGVFQIDGRSARGGCQSANDAYAEAINSGSKSSALNILREKAPKDYVLQFHNLNANLDRIFTPPLEVYVSPFCSSSFDQVPEELEEWAAENVLGAAARPLRPISIVIEGESRTGKTIWARSLGPHNYLCGHLDLSPKVYSNDAWYNVIDDVDPHYLKHFKEFMGAQRDWQSNTKYGKPVQIKGGIPAIFLCNPGPNSSYKEFLDEEKNSALKNWALKNATFVTLTGPLYSGSNQSTA |
| NCBI Accession | NP_803558.1 |
|---|---|
| Location | 2160-2453 |
| Gene Name | C4 |
| Protein Name | C4 protein |
| Coding Region | ATGGGGAACCTCATCTACACGTGCTCATCCAATTCGAGGGAAAATACCAATGCACAAACAACAGATTCTTCGACCTTATTTCCCCAACCCGGTCAGCAAATTTCCATCCGAACATTCAGAGAGCTAAAAGCTCTTCAGATGTCAAAGCCTATGTGGAGAAAGACGGAGACTTCATTGACTTTGGAGTTTTCCAAATCGATGGCAGATCAGCTAGAGGAGGTTGCCAATCTGCCAACGACGCATATGCCGAGGCAATCAATTCAGGGTCCAAGTCTTCGGCCCTCAATATTTTAA |
| Protein Sequence | MGNLIYTCSSNSRENTNAQTTDSSTLFPQPGQQISIRTFRELKALQMSKPMWRKTETSLTLEFSKSMADQLEEVANLPTTHMPRQSIQGPSLRPSIF |
References More References in PubMed
| 1 |
Nalla MK, et al. Front Plant Sci. 2023 Oct 23;14:1223982. doi: 10.3389/fpls.2023.1223982. eCollection 2023. PMID: 37936944 |
|---|---|
| 2 |
Role of Diversity and Recombination in the Emergence of Chilli Leaf Curl Virus. Mishra M, et al. Pathogens. 2022 Apr 30;11(5):529. doi: 10.3390/pathogens11050529. PMID: 35631050 |
| 3 |
Shahid MS. Virusdisease. 2020 Sep;31(3):378-382. doi: 10.1007/s13337-020-00601-2. Epub 2020 May 29. PMID: 32904994 |
| 4 |
Krishnan N, et al. J Virol Methods. 2022 Apr;302:114474. doi: 10.1016/j.jviromet.2022.114474. Epub 2022 Jan 22. PMID: 35077721 |
| 5 |
Mishra M, et al. 3 Biotech. 2020 Apr;10(4):169. doi: 10.1007/s13205-020-2159-9. Epub 2020 Mar 16. PMID: 32206503 |
| 6 |
Kushwaha NK, et al. Physiol Mol Biol Plants. 2019 Sep;25(5):1185-1196. doi: 10.1007/s12298-019-00693-1. Epub 2019 Aug 22. PMID: 31564781 |
| 7 |
Sravya G, et al. Theory Biosci. 2023 Feb;142(1):47-60. doi: 10.1007/s12064-022-00383-9. Epub 2023 Jan 6. PMID: 36607541 |
| 8 |
Chilli leaf curl disease populations in India are highly recombinant, and rapidly segregated. Pandey V, et al. 3 Biotech. 2022 Mar;12(3):83. doi: 10.1007/s13205-022-03139-w. Epub 2022 Feb 28. PMID: 35251885 |
| 9 |
Prasad KSUD, et al. Virusdisease. 2024 Sep;35(3):484-495. doi: 10.1007/s13337-024-00891-w. Epub 2024 Sep 16. PMID: 39464737 |
| 10 |
Shahid MS, et al. Virol J. 2019 Nov 9;16(1):131. doi: 10.1186/s12985-019-1235-4. PMID: 31706358 |