Tomato geminivirus 1
Basic Information
| Genus | Topilevirus |
|---|---|
| Isolate | Brazil |
| Host | |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
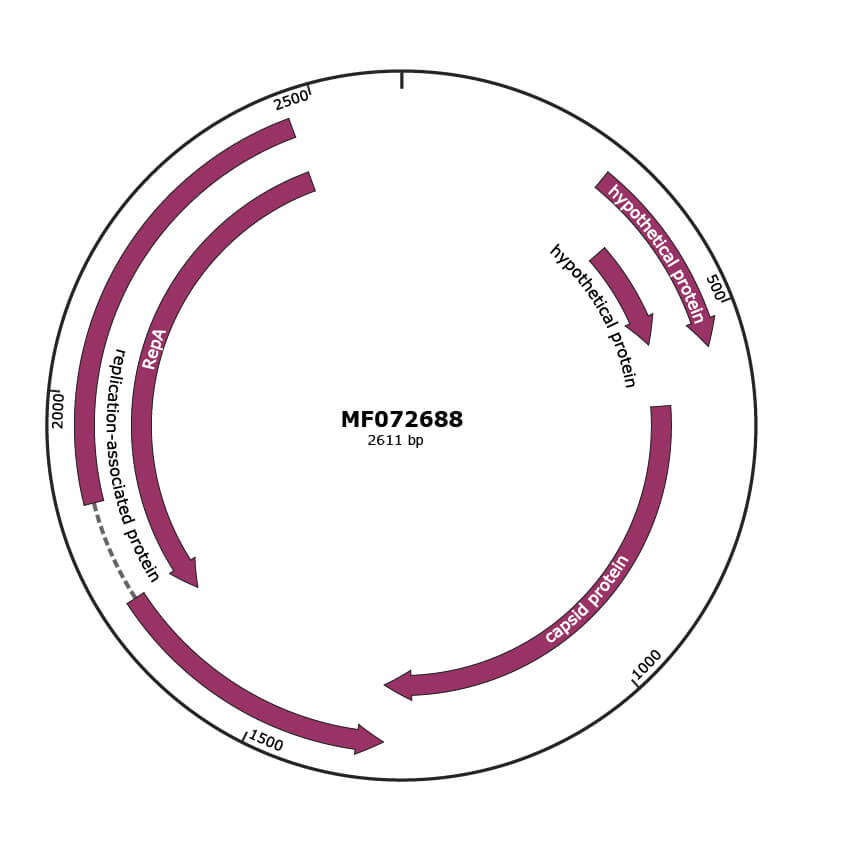
JBrowse
Genome
MF072688
Gene Information
| NCBI Accession | ASU08499.1 |
|---|---|
| Location | 285-548 |
| Protein Name | hypothetical protein |
| Coding Region | ATGGAGTGGTCTTTTAACGCAGTGGTGTACATTATTGTGTACATTTGTACGCTATTCGTTACGCTCGTTAATGGATATTCCGTTATACGAAGTGAACGACTATCCCGGCGTCTACAATCAATTTCTACAGACTTGCGCCAATCGTCTAATTTCTTGGCAATTATCGAAGGTTCAATTGGAGCTCGACGTGAAAGGAATTTCAATCGACCGGAAGGAGCAGCTTCTGAAACTCCAGTAGTTCCTCGAGGAGTTCAAGAAGTTTAA |
| Protein Sequence | MEWSFNAVVYIIVYICTLFVTLVNGYSVIRSERLSRRLQSISTDLRQSSNFLAIIEGSIGARRERNFNRPEGAASETPVVPRGVQEV |
| NCBI Accession | ASU08500.1 |
|---|---|
| Location | 355-522 |
| Protein Name | hypothetical protein |
| Coding Region | ATGGATATTCCGTTATACGAAGTGAACGACTATCCCGGCGTCTACAATCAATTTCTACAGACTTGCGCCAATCGTCTAATTTCTTGGCAATTATCGAAGGTTCAATTGGAGCTCGACGTGAAAGGAATTTCAATCGACCGGAAGGAGCAGCTTCTGAAACTCCAGTAG |
| Protein Sequence | MDIPLYEVNDYPGVYNQFLQTCANRLISWQLSKVQLELDVKGISIDRKEQLLKLQ |
| NCBI Accession | ASU08496.1 |
|---|---|
| Location | 623-1333 |
| Protein Name | capsid protein |
| Coding Region | ATGGCGGAGCGGAAGAGGAAGATGGAATGGTCGAAAGCGAATCGGAAGAGGAGTCGTACGACTCTGCCGAGGAATTCTAGGGTGTATGCTAGAAAACGACCTATATTGCGAGGTCCTAAATTGACTAGCAGGGATAAGGTTTATACGTATACGAAATACGAGACACTTACTACAGCGGGTGCGGTTTATCATCTCAATAGTTTTAGTCAGGGTTTAGCTAATAATCAACGTGCGACTTCCGTTGATATTGTTCATAATGTTCAGCTTAGGTTTTCTGTTGAGTTACCTACTGGTGCTATGGTTTTTTGTCGTCGTTACAAGATGTCATGGGCTGTTGTTCAAGATATGATGCCGGGAGATAGTATGCCAGCGGTCAGCGATATTTTTAGTATGACTTCTCCTCCTATTGAAGGTTTGGAGTATATTGCTGATGATAATCATCGCAGATTTCAAGTTAAAAAATTTGGAATTTTGTACTTGGAATCTGGTGGTGCTGACTACTCTGTTAATACTGGAAAGCAGACGTACAACGTACAAGATGCTTTGAAAAGGGTCTTCGTGAAGTGCAATACACGCGGTGTGTATGATAGTAACAGCTCTGCTGGTTCTATCAGTGCTATGAATTCTGGGGCTTTGTATTTTGTTGTAAATCCCACTTTCGCTGGGTATTTGGGTTTCAACATTACTGTGTATCATAATTCACCTGTTTAA |
| Protein Sequence | MAERKRKMEWSKANRKRSRTTLPRNSRVYARKRPILRGPKLTSRDKVYTYTKYETLTTAGAVYHLNSFSQGLANNQRATSVDIVHNVQLRFSVELPTGAMVFCRRYKMSWAVVQDMMPGDSMPAVSDIFSMTSPPIEGLEYIADDNHRRFQVKKFGILYLESGGADYSVNTGKQTYNVQDALKRVFVKCNTRGVYDSNSSAGSISAMNSGALYFVVNPTFAGYLGFNITVYHNSPV |
| NCBI Accession | ASU08498.1 |
|---|---|
| Location | NA |
| Protein Name | replication-associated protein |
| Coding Region | ATGCCTCGCCAACCCTCTTCCTTTAAAATCCAGGGCAAATCCTTTTTTCTAACCTATCCAAAGTGTCCTCTAATTCCAATTTTCATTATCGATTACTTTTTTCAATTACTAAAAGATTATAACGTCACATACGTTAGAGCTTGTACGGAAAACCATCAAGATGGAGAGCCACACATCCACTGTCTGGTACAGACTGAGAAGAGGTTTCAGACGCAGAACCAGAGGTTCTTCGACATCACCGACCCTAACGGATCGGGAATTTATCATCCAAACATCCAGATCCCAAGAAGGGATGCAGATGTTGCGGACTACATTTCCAAGGGGGGTACCTTTGAGGAGAGAGGTATTCTTAGAGCTTCCCGAAGAAGTCCGAAGAAGAATAGAGACGCTATCTGGACAAGAATTCTCTCAGAATCCACATCCAAGTCTGACTTCCTCACCAGAGTACTGGCCGAACAGCCCTACACGTATGCCAACAACTATAGAAACCTCGAATACATGGCTGATAAACAATGGCCCACACCCCCTGAGAGTTATCAACCCAGATGGACAAATTTTATTGGCATTCCAGAGTCCTTACAACACTGGGCAGATCAAAACCTCTACTGCGAACCAGAACTGGTACCAGACCGACCCACCACCGCAATAATTGAAGGTCCAACGAGATGTGGAAAAACAGCATGGGCAAGAAGCTTGGGAAAACATAATTACTACTGCGGAGGAGTCGACTGGTCAAACTATGACAACACGGCGTTATATAATGTATTCGATGATATTCCTTTCCAGTTTTTACCATGTAAAAAAGAAATCCTAGGAGCACAAAAGGATTTCACAGTAAACGAAAAATATAAGAAAAAATGTAGAATAAAGGGAGGAATACCATCTATTATCCTATGTAATTCTGATCAATCATACGAAACAGCTATACAGAATAGCGATATCTATGAATGGTCACAACAAAATGTAGAATACTTCTTTATTAATAAAAACCTGTATTAA |
| Protein Sequence | MPRQPSSFKIQGKSFFLTYPKCPLIPIFIIDYFFQLLKDYNVTYVRACTENHQDGEPHIHCLVQTEKRFQTQNQRFFDITDPNGSGIYHPNIQIPRRDADVADYISKGGTFEERGILRASRRSPKKNRDAIWTRILSESTSKSDFLTRVLAEQPYTYANNYRNLEYMADKQWPTPPESYQPRWTNFIGIPESLQHWADQNLYCEPELVPDRPTTAIIEGPTRCGKTAWARSLGKHNYYCGGVDWSNYDNTALYNVFDDIPFQFLPCKKEILGAQKDFTVNEKYKKKCRIKGGIPSIILCNSDQSYETAIQNSDIYEWSQQNVEYFFINKNLY |
| NCBI Accession | ASU08497.1 |
|---|---|
| Location | 1680-2465 |
| Protein Name | RepA |
| Coding Region | ATGCCTCGCCAACCCTCTTCCTTTAAAATCCAGGGCAAATCCTTTTTTCTAACCTATCCAAAGTGTCCTCTAATTCCAATTTTCATTATCGATTACTTTTTTCAATTACTAAAAGATTATAACGTCACATACGTTAGAGCTTGTACGGAAAACCATCAAGATGGAGAGCCACACATCCACTGTCTGGTACAGACTGAGAAGAGGTTTCAGACGCAGAACCAGAGGTTCTTCGACATCACCGACCCTAACGGATCGGGAATTTATCATCCAAACATCCAGATCCCAAGAAGGGATGCAGATGTTGCGGACTACATTTCCAAGGGGGGTACCTTTGAGGAGAGAGGTATTCTTAGAGCTTCCCGAAGAAGTCCGAAGAAGAATAGAGACGCTATCTGGACAAGAATTCTCTCAGAATCCACATCCAAGTCTGACTTCCTCACCAGAGTACTGGCCGAACAGCCCTACACGTATGCCAACAACTATAGAAACCTCGAATACATGGCTGATAAACAATGGCCCACACCCCCTGAGAGTTATCAACCCAGATGGACAAATTTTATTGGCATTCCAGAGTCCTTACAACACTGGGCAGATCAAAACCTCTACTGCGTAAGCTTCGATACCTATCTTCACATCAATCCTACTGCTGATTTTTCAGACTTGGTCTGGGCATCAGAAACAACGAAAATGGAAATTCGTGAAGCATGGGAAAACCAAGCTGAGTCACCTTCATATTTTGTTTATAGGAACCAGAACTGGTACCAGACCGACCCACCACCGCAATAA |
| Protein Sequence | MPRQPSSFKIQGKSFFLTYPKCPLIPIFIIDYFFQLLKDYNVTYVRACTENHQDGEPHIHCLVQTEKRFQTQNQRFFDITDPNGSGIYHPNIQIPRRDADVADYISKGGTFEERGILRASRRSPKKNRDAIWTRILSESTSKSDFLTRVLAEQPYTYANNYRNLEYMADKQWPTPPESYQPRWTNFIGIPESLQHWADQNLYCVSFDTYLHINPTADFSDLVWASETTKMEIREAWENQAESPSYFVYRNQNWYQTDPPPQ |
References More References in PubMed
| 1 |
Resistance gene Ty-1 restricts TYLCV infection in tomato by increasing RNA silencing. Ma X, et al. Virol J. 2024 Oct 16;21(1):256. doi: 10.1186/s12985-024-02508-6. PMID: 39415211 |
|---|---|
| 2 |
Koeda S, et al. Phytopathology. 2024 Jan;114(1):294-303. doi: 10.1094/PHYTO-04-23-0119-R. Epub 2024 Feb 12. PMID: 37321561 |
| 3 |
A novel geminivirus identified in tomato and cleome plants sampled in Brazil. Fontenele RS, et al. Virus Res. 2017 Aug 15;240:175-179. doi: 10.1016/j.virusres.2017.08.007. Epub 2017 Aug 23. PMID: 28843502 |
| 4 |
Fortes IM, et al. Phytopathology. 2023 Jul;113(7):1347-1359. doi: 10.1094/PHYTO-09-22-0334-R. Epub 2023 Aug 29. PMID: 36690608 |
| 5 |
Shen W, et al. Plant Physiol. 2018 Sep;178(1):372-389. doi: 10.1104/pp.18.00268. Epub 2018 Jul 13. PMID: 30006378 |
| 6 |
Tomato auxin biosynthesis/signaling is reprogrammed by the geminivirus to enhance its pathogenicity. Vinutha T, et al. Planta. 2020 Sep 17;252(4):51. doi: 10.1007/s00425-020-03452-9. PMID: 32940767 |
| 7 |
Nithyanandam A, et al. Mol Biol Rep. 2024 Dec 18;52(1):59. doi: 10.1007/s11033-024-10150-2. PMID: 39692918 |
| 8 |
Bian XY, et al. Phytopathology. 2007 Aug;97(8):930-7. doi: 10.1094/PHYTO-97-8-0930. PMID: 18943632 |
| 9 |
Conflon D, et al. Virus Res. 2018 Jul 15;253:124-134. doi: 10.1016/j.virusres.2018.06.003. Epub 2018 Jun 15. PMID: 29908896 |
| 10 |
A New Geminivirus Associated with a Leaf Curl Disease of Tomato in Tanzania. Chiang BT, et al. Plant Dis. 1997 Jan;81(1):111. doi: 10.1094/PDIS.1997.81.1.111B. PMID: 30870927 |