Chilli leaf curl India virus
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_002986345.1 |
| Isolate | India:North India |
| Release date | 2018/8/26 |
| Submitter | Kumar,Y., Walia,Y., Bhardwaj,P., Negi,A., Hallan,V., Zaidi,A.A. |
| Host | |
| Download | Genome |GFF3 |PEP |CDS |
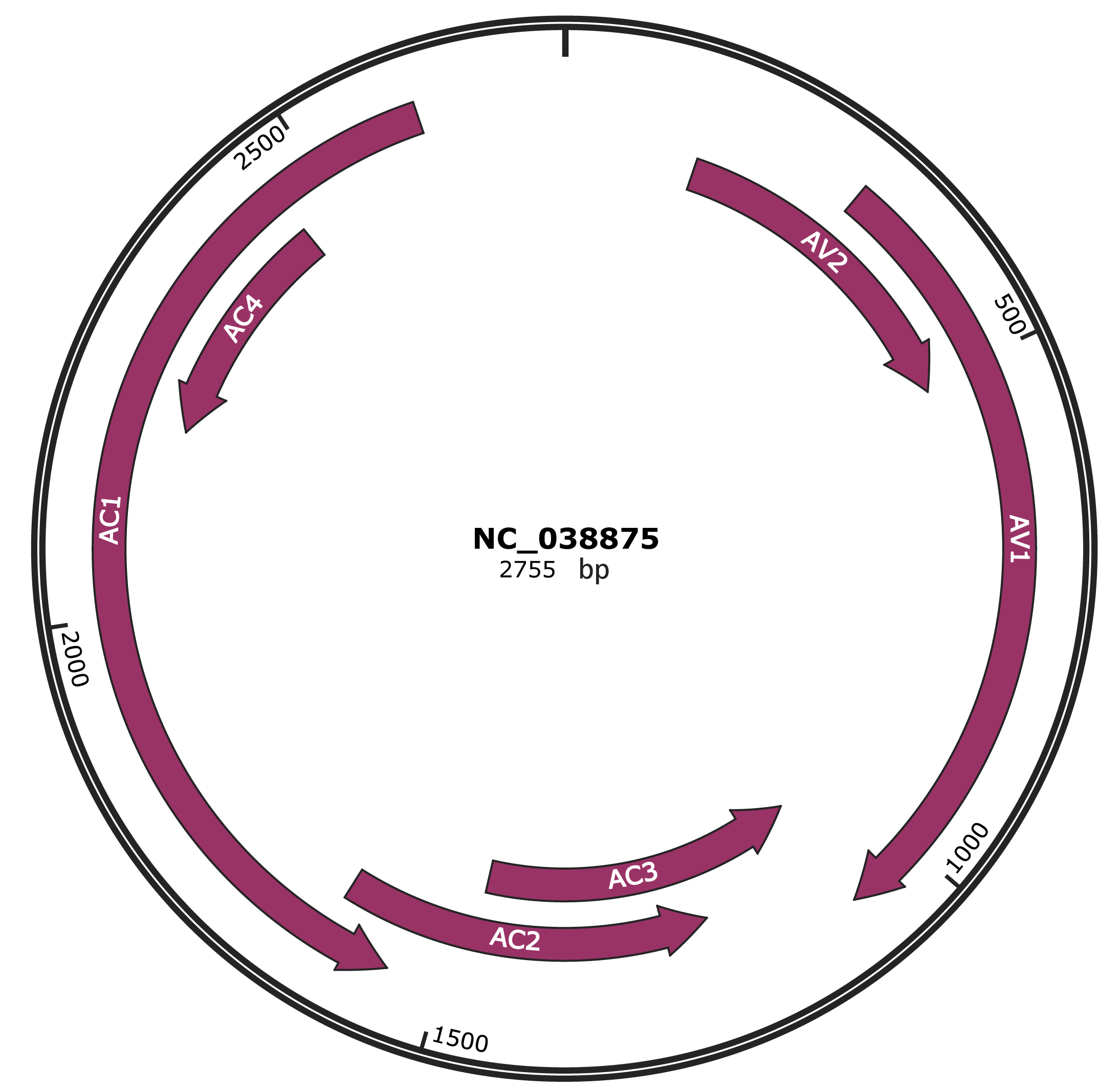
Genomic Organization
JBrowse
Genome
NC_038875
Gene Information
| NCBI Accession | YP_009508107.1 |
|---|---|
| Location | 145-510 |
| Gene Name | AV2 |
| Protein Name | pre-coat protein |
| Coding Region | ATGTGGGACCCTTTAGTAAACGAGTTTCCCGAAACCGTTCACGGTTTTAGGTGTATGCTAGCAGTTAAATACCTGCGGTTAGTAGAAAATACGTATTCCCCAGATACTCTGGGCTACGATTTAATTAGGGATTTGATTTCAGTTATTAGGGCTAGAAATTATGTCGAAGCGACCAGCAGATATAATCATTTCCACGCCCGCCTCGAAGGTACGCCGCCGTCTGAACTTCGACAGCCCATATGCGAGCCGTGCTGCTGCCCCCATTGTCCGCGTCACAAAGGCAAGAGCATGGGCGAACAGGCCCATGAACCGAAAGCCCAAGATGTACAGGGTGTACAGAAGCCCAGATGTTCCGAGGGGATGTGA |
| Protein Sequence | MWDPLVNEFPETVHGFRCMLAVKYLRLVENTYSPDTLGYDLIRDLISVIRARNYVEATSRYNHFHARLEGTPPSELRQPICEPCCCPHCPRHKGKSMGEQAHEPKAQDVQGVQKPRCSEGM |
| NCBI Accession | YP_009508108.1 |
|---|---|
| Location | 305-1075 |
| Gene Name | AV1 |
| Protein Name | coat protein |
| Coding Region | ATGTCGAAGCGACCAGCAGATATAATCATTTCCACGCCCGCCTCGAAGGTACGCCGCCGTCTGAACTTCGACAGCCCATATGCGAGCCGTGCTGCTGCCCCCATTGTCCGCGTCACAAAGGCAAGAGCATGGGCGAACAGGCCCATGAACCGAAAGCCCAAGATGTACAGGGTGTACAGAAGCCCAGATGTTCCGAGGGGATGTGAAGGCCCGTGTAAGGTCCAGTCATTTGAGTCCAGACATGATATCCAGCACATTGGTAAAGTAATGTGTGTTAGTGATGTTACTCGTGGTATTGGGCTGACTCACCGGGTTGGCAAGAGGTTCTGTGTGAAGTCCGTTTATGTTTTGGGCAAGATCTGGATGGACGAGAACATCAAGACTAAGAATCATACGAATAGTGTTATGTTTTTCCTTGTTAGAGATCGTAGGCCTGTTGATAAGCCTCAAGATTTTGGTGAGGTTTTTAACATGTTTGATAATGAGCCCAGCACGGCGACTGTGAAGAATGTTCATCGTGATAGGTATCAGGTGCTGAGAAAGTGGCATGCAACTGTCACCGGTGGTCAATATGCATCCAAGGAGCAGGCTCTCGTGAAGAAGTTTATTAGGGTTAATAATTATGTTGTGTATAACCAGCAAGAGGCTGGCAAGTATGAGAATCATACTGAGAATGCGTTGATGTTGTATATGGCGTGTACCCACGCCTCTAACCCTGTGTATGCTACATTGAAGATACGGATCTACTTCTATGATTCAGTATCGAATTAA |
| Protein Sequence | MSKRPADIIISTPASKVRRRLNFDSPYASRAAAPIVRVTKARAWANRPMNRKPKMYRVYRSPDVPRGCEGPCKVQSFESRHDIQHIGKVMCVSDVTRGIGLTHRVGKRFCVKSVYVLGKIWMDENIKTKNHTNSVMFFLVRDRRPVDKPQDFGEVFNMFDNEPSTATVKNVHRDRYQVLRKWHATVTGGQYASKEQALVKKFIRVNNYVVYNQQEAGKYENHTENALMLYMACTHASNPVYATLKIRIYFYDSVSN |
| NCBI Accession | YP_009508109.1 |
|---|---|
| Location | 1072-1476 |
| Gene Name | AC3 |
| Protein Name | replication enhancer protein |
| Coding Region | ATGGATTCACGCACAGAGGAACCCATCACTGCAGCTCAAGCAGAGAATGGCGTGTATACCTGGGAGGTTCCAAATCCCCTGTATTTCAAGATAACAGAGCACCACAACAGGCCATTCCTGATGAACAAAGACATCATAACCGTCCAAGTCCAGTTCAACCACAACCTGAGGAAAGCGTTGGGGATTCACAAATGTTTCATAGCCTTCCGAATCTGGATGACCTCACAGCCTCCGACTGGTCGTTTCTTAAGGGTCTTTAAGACCCAAGTTATTAAGTATTTAGATAATTTAGGAGTTATCAGTATTAACAATGTAGTTAGAGCAGTTGATCATGTATTATGGAATGTATTACATCATGTTGTATATGTAGAACAATCATATTCAATAAAATTTAATCTTTATTAA |
| Protein Sequence | MDSRTEEPITAAQAENGVYTWEVPNPLYFKITEHHNRPFLMNKDIITVQVQFNHNLRKALGIHKCFIAFRIWMTSQPPTGRFLRVFKTQVIKYLDNLGVISINNVVRAVDHVLWNVLHHVVYVEQSYSIKFNLY |
| NCBI Accession | YP_009508110.1 |
|---|---|
| Location | 1217-1624 |
| Gene Name | AC2 |
| Protein Name | transcriptional activator protein |
| Coding Region | ATGCAACCTTCATCACCCTCGAAGAGCCACTATACTCAGGTACCAATCAAGGTCCAACACAGAGCTGCTAAGCGTAGAGCCATCCGGCGTAAGAGGGTTGATCTAAACTGTGGGTGCTCATACTACGTACACATCGACTGCCACAAATATGGATTCACGCACAGAGGAACCCATCACTGCAGCTCAAGCAGAGAATGGCGTGTATACCTGGGAGGTTCCAAATCCCCTGTATTTCAAGATAACAGAGCACCACAACAGGCCATTCCTGATGAACAAAGACATCATAACCGTCCAAGTCCAGTTCAACCACAACCTGAGGAAAGCGTTGGGGATTCACAAATGTTTCATAGCCTTCCGAATCTGGATGACCTCACAGCCTCCGACTGGTCGTTTCTTAAGGGTCTTTAA |
| Protein Sequence | MQPSSPSKSHYTQVPIKVQHRAAKRRAIRRKRVDLNCGCSYYVHIDCHKYGFTHRGTHHCSSSREWRVYLGGSKSPVFQDNRAPQQAIPDEQRHHNRPSPVQPQPEESVGDSQMFHSLPNLDDLTASDWSFLKGL |
| NCBI Accession | YP_009508111.1 |
|---|---|
| Location | 1554-2612 |
| Gene Name | AC1 |
| Protein Name | replication initiator protein |
| Coding Region | ATGCCTCGTGCTCATCAGTTCCAAGTTAAAGCCAAAAATATCTTCCTTACTTATCCAAAATGCCCAATACCCAAAGAGCAAATGCTCGAACTCCTAAAAAACGTTTCTTGTCCTTCTGATAAATTATTTATCAGAGTGTCACAGGAAAAACACCAAGATGGGTCTCTGCATATCCATGCCCTCATCCAATTCAAAGGTAAATCCCAATTCAGAAACCCCAGACATTTCGATGTCACTCACCCTAATACATCAACCCAATTCCACCCAAACTTCCAGGGAGCAAAGTCCAGCTCCGATGTCAAGTCCTATATCGAGAAGGACGGTGATTACATCGACTGGGGTCAGTTTCAGGTCGATGGACGATCTGCTCGAGGAGGTCAACAGACAGCTAATGATGCTGCAGCAGAGGCCCTAAATGCAGGTTCAGCTGAAGCAGCTCTAGCAATTATTAGGGAGAAACTCCCGAAAGATTTTATTTTTCAATATCATAATTTAAAAACTAATTTAGATAGGATTTTTACACCTCCAGTGGAGGTTTATGTTCCCCCATTTCTTTCTTCTTCGTTCGATCAGGTTCCAGAGGCCATAGAGGAATGGGCCTCTGAAAATGTGATGGGTCCCGCTGCGCGGCCATTGAGACCTAAAAGTATCGTCATTGAGGGTGATAGTCGTACAGGCAAGACAATGTGGGCCAGGTCTTTGGGACCACACAATTATTTATGCGGCCATTTAGATCTGAGTCCCAAGATATATTCAAATGATGCCTGGTACAACGTCATTGATGACGTAGATCCCCACTACCTAAAGCACTTTAAAGAGTTCATGGGGGCCCAAAGGGACTGGCAAAGCAACACCAAGTACGGGAAGCCAGTTCAAATTAAAGGGGGCATTCCCACTATCTTCCTCTGCAATCCTGGGCCCAATTCCAGCTATAAAGAATTCCTCGATGAGGAAAAGAACTCAGCACTGAGGTCGTGGGCAATACATAATGCAACCTTCATCACCCTCGAAGAGCCACTATACTCAGGTACCAATCAAGGTCCAACACAGAGCTGCTAA |
| Protein Sequence | MPRAHQFQVKAKNIFLTYPKCPIPKEQMLELLKNVSCPSDKLFIRVSQEKHQDGSLHIHALIQFKGKSQFRNPRHFDVTHPNTSTQFHPNFQGAKSSSDVKSYIEKDGDYIDWGQFQVDGRSARGGQQTANDAAAEALNAGSAEAALAIIREKLPKDFIFQYHNLKTNLDRIFTPPVEVYVPPFLSSSFDQVPEAIEEWASENVMGPAARPLRPKSIVIEGDSRTGKTMWARSLGPHNYLCGHLDLSPKIYSNDAWYNVIDDVDPHYLKHFKEFMGAQRDWQSNTKYGKPVQIKGGIPTIFLCNPGPNSSYKEFLDEEKNSALRSWAIHNATFITLEEPLYSGTNQGPTQSC |
| NCBI Accession | YP_009508112.1 |
|---|---|
| Location | 2198-2455 |
| Gene Name | AC4 |
| Protein Name | AC4 protein |
| Coding Region | ATGGGTCTCTGCATATCCATGCCCTCATCCAATTCAAAGGTAAATCCCAATTCAGAAACCCCAGACATTTCGATGTCACTCACCCTAATACATCAACCCAATTCCACCCAAACTTCCAGGGAGCAAAGTCCAGCTCCGATGTCAAGTCCTATATCGAGAAGGACGGTGATTACATCGACTGGGGTCAGTTTCAGGTCGATGGACGATCTGCTCGAGGAGGTCAACAGACAGCTAATGATGCTGCAGCAGAGGCCCTAA |
| Protein Sequence | MGLCISMPSSNSKVNPNSETPDISMSLTLIHQPNSTQTSREQSPAPMSSPISRRTVITSTGVSFRSMDDLLEEVNRQLMMLQQRP |
References More References in PubMed
| 1 |
Nalla MK, et al. Front Plant Sci. 2023 Oct 23;14:1223982. doi: 10.3389/fpls.2023.1223982. eCollection 2023. PMID: 37936944 |
|---|---|
| 2 |
Chilli leaf curl disease populations in India are highly recombinant, and rapidly segregated. Pandey V, et al. 3 Biotech. 2022 Mar;12(3):83. doi: 10.1007/s13205-022-03139-w. Epub 2022 Feb 28. PMID: 35251885 |
| 3 |
Role of Diversity and Recombination in the Emergence of Chilli Leaf Curl Virus. Mishra M, et al. Pathogens. 2022 Apr 30;11(5):529. doi: 10.3390/pathogens11050529. PMID: 35631050 |
| 4 |
Krishnan N, et al. J Virol Methods. 2022 Apr;302:114474. doi: 10.1016/j.jviromet.2022.114474. Epub 2022 Jan 22. PMID: 35077721 |
| 5 |
Prasad KSUD, et al. Virusdisease. 2024 Sep;35(3):484-495. doi: 10.1007/s13337-024-00891-w. Epub 2024 Sep 16. PMID: 39464737 |
| 6 |
Sravya G, et al. Theory Biosci. 2023 Feb;142(1):47-60. doi: 10.1007/s12064-022-00383-9. Epub 2023 Jan 6. PMID: 36607541 |
| 7 |
First Report of Chilli leaf curl India virus Infecting Mentha spicata (Neera) in India. Saeed ST, et al. Plant Dis. 2014 Jan;98(1):164. doi: 10.1094/PDIS-07-13-0750-PDN. PMID: 30708610 |
| 8 |
Mishra M, et al. 3 Biotech. 2020 Apr;10(4):169. doi: 10.1007/s13205-020-2159-9. Epub 2020 Mar 16. PMID: 32206503 |
| 9 |
Krishnan N, et al. Physiol Mol Biol Plants. 2025 Aug;31(8):1363-1374. doi: 10.1007/s12298-025-01570-w. Epub 2025 Mar 7. PMID: 41080515 |
| 10 |
Kushwaha NK, et al. Physiol Mol Biol Plants. 2019 Sep;25(5):1185-1196. doi: 10.1007/s12298-019-00693-1. Epub 2019 Aug 22. PMID: 31564781 |