Beet curly top virus
Basic Information
| Genus | Curtovirus |
|---|---|
| NCBI Assembly | GCF_000843505.1 |
| Release date | 2018/11/26 |
| Submitter | Stanley,J., Markham,P.G., Callis,R.J., Pinner,M.S. |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
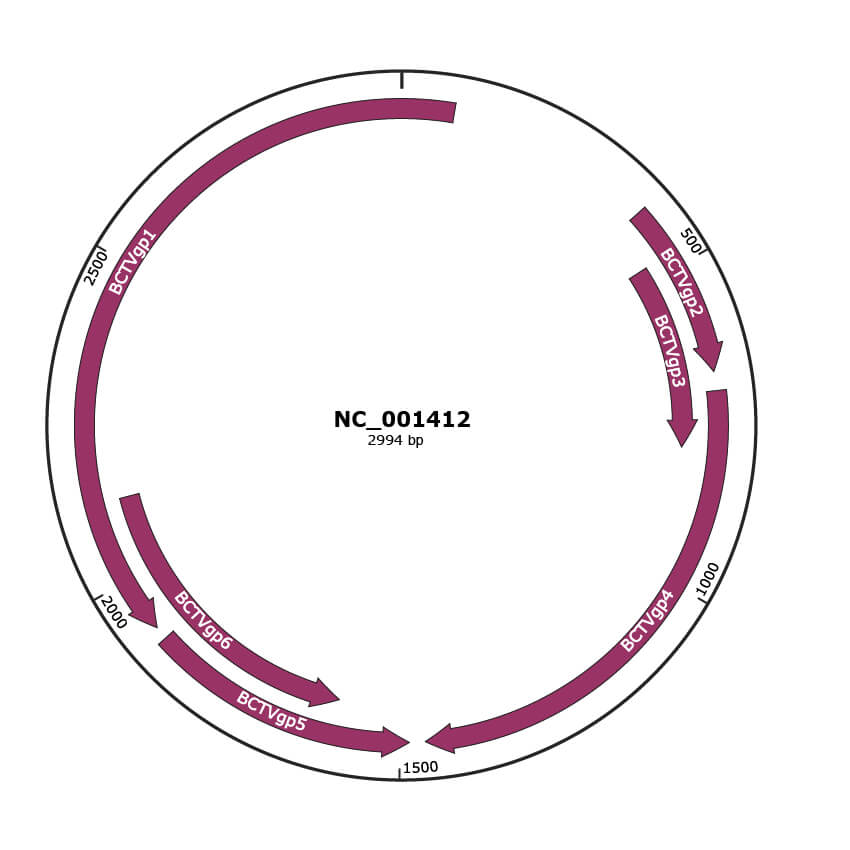
JBrowse
Genome
NC_001412
Gene Information
| NCBI Accession | NP_040557.1 |
|---|---|
| Location | 1917-2994,1-80 |
| Protein Name | replication-associated protein |
| Coding Region | ATGAAAGTTGTACTAATATATATAATACTCCAATACTCCAATAGATATGTTAATATGCTAAGCCACGTCATCGGAGTACANATGCCTCCTACTAAAAGATTTCGTATTCAAGCAAAAAACATATTTCTTACATATCCTCAGTGTTCTCTTTCAAAAGAAGAAGCTCTTGAGCAAATTCAAAGAATACAACTTTCATCTAATAAAAAATATATTAAAATTGCCAGAGAGCTACACGAAGATGGGCAACCTCATCTCCACGTCCTGCTTCAACTCGAAGGAAAAGTTCAGATCACAAATATCAGATTATTCGACCTGGTATCCCCAACCAGGTCAGCACATTTCCATCCAAACATTCAGAGAGCTAAATCCAGCTCCGACGTCAAGTCCTACGTCGACAAGGACGGAGACACAATTGAATGGGGAGAATTCCAGATCGACGGTAGAAGTGCTAGAGGAGGTCAACAGACAGCTAACGACTCATATGCCAAGGCGTTAAACGCAACTTCTCTTGACCAAGCACTTCAAATATTGAAGGAAGAACAACCAAAGGATTACTTCCTTCAACATCACAATCTTTTGAACAATGCTCAAAAGATATTTCAGAGGCCACCTGATCCATGGACTCCACTATTTCCTCTGTCCTCATTCACAAACGTTCCTGAGGAAATGCAAGAATGGGCTGATGCATATTTCGGGGTTGATGCCGCTGCGCGGCCTTTAAGATATAATAGTATCATAGTAGAGGGTGATTCAAGAACAGGGAAGACTATGTGGGCTAGATCTTTAGGGGCCCACAATTACATCACAGGGCACTTAGATTTTAGCCCTAGAACGTATTATGATGAAGTGGAATACAACGTCATTGATGACGTAGATCCCACTTACTTAAAGATGAAACACTGGAAACACCTTATTGGAGCACAAAAGGAGTGGCAGACAAACTTAAAGTATGGAAAACCACGTGTCATTAAAGGTGGTATCCCCTGCATTATATTATGCAATCCAGGACCTGAGAGCTCATACCAACAATTTCTTGAAAAACCAGAAAATGAAGCCCTTAAGTCCTGGACATTACATAATTCAACCTTCTGCAAACTCCAAGGTCCGCTCTTTAATAACCAAGCAGCAGCATCCTCGCAAGGTGACTCTACCCTGTAA |
| Protein Sequence | MKVVLIYIILQYSNRYVNMLSHVIGVXMPPTKRFRIQAKNIFLTYPQCSLSKEEALEQIQRIQLSSNKKYIKIARELHEDGQPHLHVLLQLEGKVQITNIRLFDLVSPTRSAHFHPNIQRAKSSSDVKSYVDKDGDTIEWGEFQIDGRSARGGQQTANDSYAKALNATSLDQALQILKEEQPKDYFLQHHNLLNNAQKIFQRPPDPWTPLFPLSSFTNVPEEMQEWADAYFGVDAAARPLRYNSIIVEGDSRTGKTMWARSLGAHNYITGHLDFSPRTYYDEVEYNVIDDVDPTYLKMKHWKHLIGAQKEWQTNLKYGKPRVIKGGIPCIILCNPGPESSYQQFLEKPENEALKSWTLHNSTFCKLQGPLFNNQAAASSQGDSTL |
| NCBI Accession | NP_899663.1 |
|---|---|
| Location | 401-667 |
| Protein Name | movement protein |
| Coding Region | ATGATGGTCTGTCTACCAGACTGGTTATTTTTGCTATTTATCTTCAGTATTCTACTGCAATCAGGTACGAACTTTTATGGGACCTTTCAGAGTGGATCAATTTCCAGACAATTATCCAGCCTTTCTAGCAGTATCGACCAGTTGTTTCTTAAGGTACAACAGGTGGTGTATACTAGGTATCCATCAAGAGATAGGGTCTCTGACCCTAGAAGAAGGCGAGGTCTTTCGTCAATTCCAGAAGGAAGTGAAGAAGCTACTGAGGTGTAA |
| Protein Sequence | MMVCLPDWLFLLFIFSILLQSGTNFYGTFQSGSISRQLSSLSSSIDQLFLKVQQVVYTRYPSRDRVSDPRRRRGLSSIPEGSEEATEV |
| NCBI Accession | NP_040558.1 |
|---|---|
| Location | 477-785 |
| Protein Name | hypothetical protein |
| Coding Region | ATGGGACCTTTCAGAGTGGATCAATTTCCAGACAATTATCCAGCCTTTCTAGCAGTATCGACCAGTTGTTTCTTAAGGTACAACAGGTGGTGTATACTAGGTATCCATCAAGAGATAGGGTCTCTGACCCTAGAAGAAGGCGAGGTCTTTCGTCAATTCCAGAAGGAAGTGAAGAAGCTACTGAGGTGTAAGGTTAATTTTCATAGGAAGTGTTCGTTGTATGAGGAAATATACAAGAAATACGTATACAATGTCCCAGAAAAGAAAGGTGAATCCTCAAAGTGCGTGGCCGAAGAAGAGGAGGACTAG |
| Protein Sequence | MGPFRVDQFPDNYPAFLAVSTSCFLRYNRWCILGIHQEIGSLTLEEGEVFRQFQKEVKKLLRCKVNFHRKCSLYEEIYKKYVYNVPEKKGESSKCVAEEEED |
| NCBI Accession | NP_040559.1 |
|---|---|
| Location | 697-1461 |
| Protein Name | coat protein |
| Coding Region | ATGAGGAAATATACAAGAAATACGTATACAATGTCCCAGAAAAGAAAGGTGAATCCTCAAAGTGCGTGGCCGAAGAAGAGGAGGACTAGTACGACTTCGAGGAAATACCAATGGAGGAGACCTGTGACAAAAAACAGGACTCTGAAGTTAAAGATGTATGATGATATGTTGGGTGCTGGTGGTATAGGATCTACCATTAGTAATAATGGTATGATTACTATGTTGAATAATTATGTCCAGGGTATTGGTGATAGTCAGAGAGCGAAGAACGTTACTGTGACGAAGCATTTGAAGTTTGATATGGCTCTTATGGGGAGTTCTCAGTTCTGGGAGACTCCTAATTATATGACCCAATATCATTGGATTATCATAGACAAGGATGTTGGGTCAGTGTTTCCTACTAAGTTATCGAGTATATTTGATATTCCCGATAACGGTCAGGCTATGCCGTCTACTTATCGTATTCGAAGAGATATGAACGAGAGGTTTATTGGGAAGAAGAAATGGAGAACTCATTGGATGTCTACTGGTACTGGATATGGAGGGAAGGAGACTTACAAGGCTCCTTCAATGCCAAATTACAAGAAACCGATGAATATCAATGTGCGCAATTTGAACATGAGGACTATTTGGAAAGACACCGGTGGTGGGAAGTATGAAGACGTGAAGGAGAATGCTTTACTCTATGTTGTTGTTAATGATAATACGGATAATACTAATATGTATGCCACATTATTTGGCAATTGTAGATGCTATTTTTATTAA |
| Protein Sequence | MRKYTRNTYTMSQKRKVNPQSAWPKKRRTSTTSRKYQWRRPVTKNRTLKLKMYDDMLGAGGIGSTISNNGMITMLNNYVQGIGDSQRAKNVTVTKHLKFDMALMGSSQFWETPNYMTQYHWIIIDKDVGSVFPTKLSSIFDIPDNGQAMPSTYRIRRDMNERFIGKKKWRTHWMSTGTGYGGKETYKAPSMPNYKKPMNINVRNLNMRTIWKDTGGGKYEDVKENALLYVVVNDNTDNTNMYATLFGNCRCYFY |
| NCBI Accession | NP_040560.1 |
|---|---|
| Location | 1486-1896 |
| Protein Name | replication enhancer protein |
| Coding Region | ATGAATGTAATAGAGGATTTTCGCACAGGGGAACCTATTACTCTCCATCAGGCAACAAATTCCGTCGAATTCGAGAATGTACCGAATCCACTGTATATGAAACTCCTATGGTTCGAGAGATACGGGCCAATCTATCAACTGAAGATACAAATCAGATTCAACTACAACCTCCGGAGAGCGTTGAATCTTCACAAGTGCTGGATAGAGCTGACGATAACTGGATCGAACAGGATATTGACTGGACCCCGTTTCTTGAAGGTCTTGAAAAAGAGACTAGAGATATACTTGGATAATCTAGGTTTAATTTGTATTAATAATGTAATTAGAGGTTTAAATCATGTCCTGTATGAAGAATTTACTTTTGTATCAAGTGTAATTCAGAACCAGAGTGTTGCAATGAACTTGTACTAA |
| Protein Sequence | MNVIEDFRTGEPITLHQATNSVEFENVPNPLYMKLLWFERYGPIYQLKIQIRFNYNLRRALNLHKCWIELTITGSNRILTGPRFLKVLKKRLEIYLDNLGLICINNVIRGLNHVLYEEFTFVSSVIQNQSVAMNLY |
| NCBI Accession | NP_040561.1 |
|---|---|
| Location | 1604-2125 |
| Protein Name | pathogenicity determinant protein |
| Coding Region | ATGGAAAACCACGTGTCATTAAAGGTGGTATCCCCTGCATTATATTATGCAATCCAGGACCTGAGAGCTCATACCAACAATTTCTTGAAAAACCAGAAAATGAAGCCCTTAAGTCCTGGACATTACATAATTCAACCTTCTGCAAACTCCAAGGTCCGCTCTTTAATAACCAAGCAGCAGCATCCTCGCAAGGTGACTCTACCCTGTAACTGCCACTTCACAATACACCATGAATGTAATAGAGGATTTTCGCACAGGGGAACCTATTACTCTCCATCAGGCAACAAATTCCGTCGAATTCGAGAATGTACCGAATCCACTGTATATGAAACTCCTATGGTTCGAGAGATACGGGCCAATCTATCAACTGAAGATACAAATCAGATTCAACTACAACCTCCGGAGAGCGTTGAATCTTCACAAGTGCTGGATAGAGCTGACGATAACTGGATCGAACAGGATATTGACTGGACCCCGTTTCTTGAAGGTCTTGAAAAAGAGACTAGAGATATACTTGGATAA |
| Protein Sequence | MENHVSLKVVSPALYYAIQDLRAHTNNFLKNQKMKPLSPGHYIIQPSANSKVRSLITKQQHPRKVTLPCNCHFTIHHECNRGFSHRGTYYSPSGNKFRRIRECTESTVYETPMVREIRANLSTEDTNQIQLQPPESVESSQVLDRADDNWIEQDIDWTPFLEGLEKETRDILG |
References More References in PubMed
| 1 |
Han J, et al. J Gen Virol. 2024 Jul;105(7):002012. doi: 10.1099/jgv.0.002012. PMID: 39073409 |
|---|---|
| 2 |
Complementary-sense gene regulation in beet curly top virus-SpCT. Guerrero J, et al. Arch Virol. 2019 Nov;164(11):2823-2828. doi: 10.1007/s00705-019-04385-9. Epub 2019 Sep 4. PMID: 31485748 |
| 3 |
Alkhatib BM, et al. Viruses. 2024 Oct 5;16(10):1571. doi: 10.3390/v16101571. PMID: 39459905 |
| 4 |
Ebrahimi S, et al. Viruses. 2023 Sep 26;15(10):1996. doi: 10.3390/v15101996. PMID: 37896771 |
| 5 |
Regulatory Roles of Small Non-coding RNAs in Sugar Beet Resistance Against Beet curly top virus. Majumdar R, et al. Front Plant Sci. 2022 Jan 10;12:780877. doi: 10.3389/fpls.2021.780877. eCollection 2021. PMID: 35082811 |
| 6 |
Withycombe J, et al. BMC Genomics. 2024 Dec 23;25(1):1237. doi: 10.1186/s12864-024-11143-y. PMID: 39716086 |
| 7 |
Melgarejo TA, et al. Virology. 2024 Mar;591:109981. doi: 10.1016/j.virol.2024.109981. Epub 2024 Jan 4. PMID: 38211381 |
| 8 |
Strausbaugh CA, et al. Plant Dis. 2017 Aug;101(8):1373-1382. doi: 10.1094/PDIS-03-17-0381-RE. Epub 2017 May 25. PMID: 30678603 |
| 9 |
Pest categorisation of Beet curly top virus (non-EU isolates). EFSA Panel on Plant Health (PLH), et al. EFSA J. 2017 Oct 11;15(10):e04998. doi: 10.2903/j.efsa.2017.4998. eCollection 2017 Oct. PMID: 32625295 |
| 10 |
Melgarejo TA, et al. Plant Dis. 2022 Dec;106(12):3022-3026. doi: 10.1094/PDIS-04-22-0856-SC. Epub 2022 Nov 17. PMID: 35549320 |