Citrus chlorotic dwarf associated virus
Basic Information
| Genus | Citlodavirus |
|---|---|
| NCBI Assembly | GCF_000898955.1 |
| Isolate | Turkey |
| Release date | 2015/2/22 |
| Submitter | Loconsole,G., Saldarelli,P., Doddapaneni,H., Savino,V., Martelli,G.P., Saponari,M. |
| Host | |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
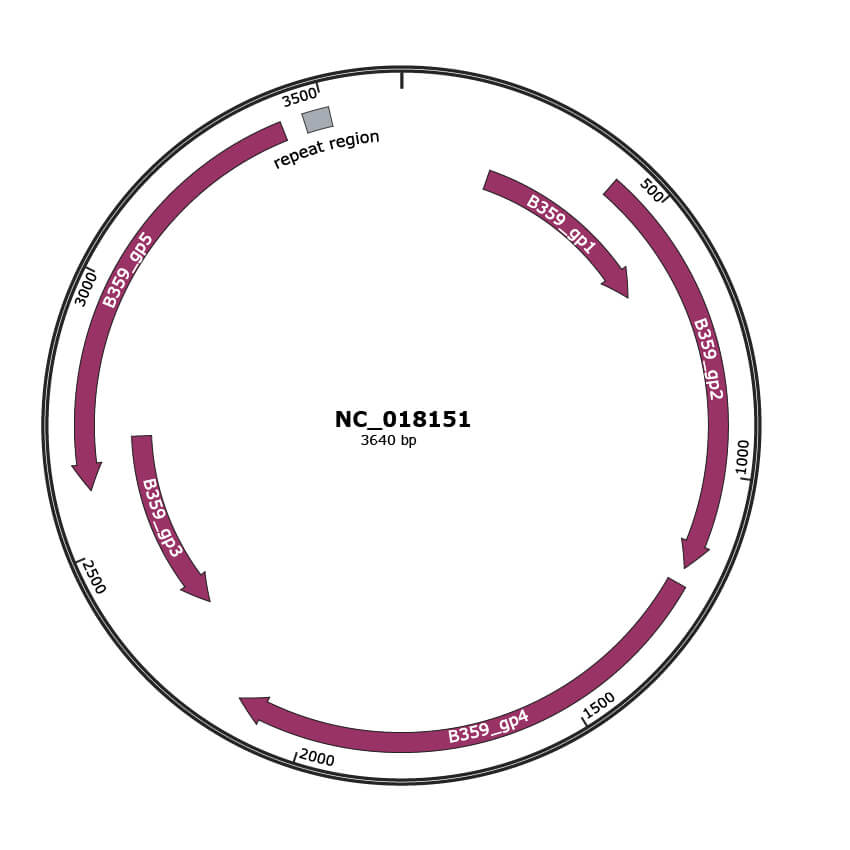
JBrowse
Genome
NC_018151
Gene Information
| NCBI Accession | YP_006522418.1 |
|---|---|
| Location | 194-613 |
| Protein Name | putative V2-like protein |
| Coding Region | ATGTGTCATTATGCATTAAGTGTTCAAGATTTGCCCGAGAGTTTGTTCGGTTTAATGAGCATGTTGAGTGTGAGGTATTTAAAGTGTGTGGAAGAGAGGGAAATGGAGAGAAGCCTAATGGTAGGGAATCCCGTAGGAGGAGCCCTGGGTAATGCTCGTATACTTATACGGTTAATACGTAGATACTGCAGGTGTAGAGACTGGGTAAGGAAAGGTAGGGTCAATGGTGAGTACCAGGAGTGGAGGACAATATGGGACAAGCCGTGTAACGACTGTGGAGACGCTGCCGATGTGGGCCACAAAGAGAAGAAGGAGGCCCAGGTGGCCGAGAAAGGAAAAGAAGCCCAAGATTGCTGGGGCTGTATTTGTGAAGGCCCGGCCCAGGAGAAAGTCGAGCAAAGGTGTTCCTCCGGGGTGTAA |
| Protein Sequence | MCHYALSVQDLPESLFGLMSMLSVRYLKCVEEREMERSLMVGNPVGGALGNARILIRLIRRYCRCRDWVRKGRVNGEYQEWRTIWDKPCNDCGDAADVGHKEKKEAQVAEKGKEAQDCWGCICEGPAQEKVEQRCSSGV |
| NCBI Accession | YP_006522419.1 |
|---|---|
| Location | 417-1181 |
| Protein Name | putative coat protein-like protein |
| Coding Region | ATGGTGAGTACCAGGAGTGGAGGACAATATGGGACAAGCCGTGTAACGACTGTGGAGACGCTGCCGATGTGGGCCACAAAGAGAAGAAGGAGGCCCAGGTGGCCGAGAAAGGAAAAGAAGCCCAAGATTGCTGGGGCTGTATTTGTGAAGGCCCGGCCCAGGAGAAAGTCGAGCAAAGGTGTTCCTCCGGGGTGTAAGGGCCCATGTAAAACACACACGGTGGATGTGATAAAGACTATTTATCACGATGGCCGTGGATCTGGAATGATTTCAAATATTGACCGAGGTGATGAGCTAGGTCAAAGGGAAGGAAGGAAAATAAGGGTTTCACGTATGATCATACGTGGCAAGATCTGGTTGGACGTGAATAATGCATCCGTACCAGGAAGCAATTTAGCTAAAATATGGATTTTCAAGGATAGGAGACCGGGGACTGAACCGGTTGCTTTTAATGCGCTGATGGATATGTCTGATTCAGAACCACTCAGTGCATTTGTGAAGGTTGACTACAGGGACAGGTTTATTGCCCTTCACACTATGACGGTAGACCTGCACGGTGGAAAGGATTTTAGGGTTGACGAGCTTGACTTGGATGAGTTAGTTGAGATAAACAGTGATGTTTTGTTTAGTCATGAGGACGATGGGTCTGTGGCCCATACGATCCAAAATGGTATTTTTATATATTATGCTTGTAGTGATCCTAGACAGACTGTGCAAATAACTGCACAGGCTCGATTGTATTTTTATGATTCTACATCAAATTAA |
| Protein Sequence | MVSTRSGGQYGTSRVTTVETLPMWATKRRRRPRWPRKEKKPKIAGAVFVKARPRRKSSKGVPPGCKGPCKTHTVDVIKTIYHDGRGSGMISNIDRGDELGQREGRKIRVSRMIIRGKIWLDVNNASVPGSNLAKIWIFKDRRPGTEPVAFNALMDMSDSEPLSAFVKVDYRDRFIALHTMTVDLHGGKDFRVDELDLDELVEINSDVLFSHEDDGSVAHTIQNGIFIYYACSDPRQTVQITAQARLYFYDSTSN |
| NCBI Accession | YP_006522420.1 |
|---|---|
| Location | 1211-2131 |
| Protein Name | putative BL1-like movement protein |
| Coding Region | ATGGACGGTCAAGATTTGGTGTTACAAGACTATCATAGCACGAGACGTATTGAATACCCTCTAAGTGATGAGTGGCAGCAGATAAAGCTTGCATTCCCTAGTATGAAGGAGATTAGCTGGCATAAGTTACGTGGTCAATGCATGAAAATTGACCATTGTCAGATACGGTATGATCCGCAAGTACCTGCTAATGCAGAAGGGAATGTATTGGTTGTGGTACACGATAGACGTATGGAAGCTGACAAGTCAATGCAAGCTGAATATACTTTTCCAATACGATGTGGAATAGAACTTAATTACTATTCCTGTTCGTATTTTTCGTTGAAGGACCCAGTACCATGGTGTGTATACTATAGGGTTGTGAACTCTACTGTGTTGAAGGGTTCTCATTTTTGTCAATTTAAGGCGCGCGTGAAGCTAAGCGCGGCTAAATCAAGTAGCCCAATTGGGTTCAGGGGTCCAAGTGTTAAGATTTTAAACAAGGCATTCAACGAAGACCAGGTGGATTTCATGCACGTGGGCATCCCGAAGTCAGAAAGGGTGTTATGCAGGAGTAATAGTGTTTTAACGACCCGGCCCAGATTGAATCTTGAGGCTGGGGAGAGTTGGGCCTCGAAAAGTATATTATCAGGCGAAGGTGGATCGGAGGTTGGTGATTCCGGCCCATATAGGGGTTTGGCTCAGTTAGGCCCAGACGCGATTGATCCAGGTGATAGCGCGTCTAATTTGGGTGATCCAAAATCAGTTGCGGATGAAGTCATTAGAAGACTTAACAGTTCAGTTATTGGGATGGACTTAAACAGTTCAAAATTTGCAGAGATAATTGGAGATGCAGTACTCAAGGGAAGTGTGATCAACAGTAGAGATAATCAGGCCTCAACAAGTAATGCTAATGATTATAAGAAAAAAAGCTTGGCTTAA |
| Protein Sequence | MDGQDLVLQDYHSTRRIEYPLSDEWQQIKLAFPSMKEISWHKLRGQCMKIDHCQIRYDPQVPANAEGNVLVVVHDRRMEADKSMQAEYTFPIRCGIELNYYSCSYFSLKDPVPWCVYYRVVNSTVLKGSHFCQFKARVKLSAAKSSSPIGFRGPSVKILNKAFNEDQVDFMHVGIPKSERVLCRSNSVLTTRPRLNLEAGESWASKSILSGEGGSEVGDSGPYRGLAQLGPDAIDPGDSASNLGDPKSVADEVIRRLNSSVIGMDLNSSKFAEIIGDAVLKGSVINSRDNQASTSNANDYKKKSLA |
| NCBI Accession | YP_006522421.1 |
|---|---|
| Location | 2300->2707 |
| Protein Name | putative C1:C2-like protein |
| Coding Region | CGAGCACAGGAACCAGACTCCAAGCCCGACAGGCCCAAATCCCTCTACATCTGCGGCCCAAGCCGATCAGGCAAAACCGCCTGGGCCAGAAGTTTAGGCCTACACAACTACTTCACGGGTGCCATCAAATTCCACGACTACAACGACCACGCACTGTATAACGTCATTGATGACATCCAATACACCAAAATCTCACATGAGGTTATGAAATCCTTAGTGGGGTCCCAAAAAAACATTACCGTTAACATTAAGTATAGACCCGATCGCACTATTAAGGGAGGCATACCGTCCATAATATGTGTTAATCCAGATATGGACTGGTTAACTTATATGTCACCCACAATAAAGGACTGGTGGAACCAAAATGTACTTATGCATTATATGGACCCCACTGATGTATTCTACTAG |
| Protein Sequence | RAQEPDSKPDRPKSLYICGPSRSGKTAWARSLGLHNYFTGAIKFHDYNDHALYNVIDDIQYTKISHEVMKSLVGSQKNITVNIKYRPDRTIKGGIPSIICVNPDMDWLTYMSPTIKDWWNQNVLMHYMDPTDVFY |
| NCBI Accession | YP_006522422.1 |
|---|---|
| Location | 2611-3420 |
| Protein Name | putative RepA-like protein |
| Coding Region | ATGGCTTCCACTTCCTCTAGCTTCCGATTCTCAGCCAAAAATATTTTCCTTACATACCCCAAGTGCCCATGCACCAAGGAACACCTCCAAGCTTTCCTTCGACTAACACTCGCACGATTCACAATCACTTACATGTGCGTGTGTGAGGAACTCCACGAATCCGGAGACCCACACCTTCATGCCATGATCCAGTGCAAGAAACGGGTCGAAACACAGAACCCCCGGTTCTTCGACCTCCTCTCCGTACGCAGAGAGAGGTCATTCCATCCCTGTATCGAGTCCTTGAAATCACCAGCCGCCTCCAGAAAATACCTCATGAAGGACGGCAACTATGTGGAGGAAGGACGCTTCAACTCCAGAGCACGATCCCCACAAAAGGATCAAGAAAAACTGTGGAGGGACGTGCTCCTCGAGGCAACCGACGAACGGTCCTTCCTTAACCTAGTCAGAGAGCTCAGACCATCAGACTTCGTCCTCAGATGGCCAGCGATCTCAGCGTTCGCACGTGACAACTACTGCAGACTACGTGAACCGTTCATACCTGCTTTCACAGAATTCCCGAACCTTCCTGAACACGTCAAGCAGTGGGCCCAGCAGAACATCCTATGTGTAAGTAAACCATTTTTACAGTATGAACTGTGTTATTCCTGCTGTCCAAAAGCAATATTTGAGACTGAGTGCCCAATTAATTTGCAACATCACTTCTGGTGTGACGAGCACAGGAACCAGACTCCAAGCCCGACAGGCCCAAATCCCTCTACATCTGCGGCCCAAGCCGATCAGGCAAAACCGCCTGGGCCAGAAGTTTAG |
| Protein Sequence | MASTSSSFRFSAKNIFLTYPKCPCTKEHLQAFLRLTLARFTITYMCVCEELHESGDPHLHAMIQCKKRVETQNPRFFDLLSVRRERSFHPCIESLKSPAASRKYLMKDGNYVEEGRFNSRARSPQKDQEKLWRDVLLEATDERSFLNLVRELRPSDFVLRWPAISAFARDNYCRLREPFIPAFTEFPNLPEHVKQWAQQNILCVSKPFLQYELCYSCCPKAIFETECPINLQHHFWCDEHRNQTPSPTGPNPSTSAAQADQAKPPGPEV |
References More References in PubMed
| 1 |
Zhao J, et al. Plant Dis. 2024 Nov;108(11):3393-3399. doi: 10.1094/PDIS-12-23-2575-RE. Epub 2024 Nov 6. PMID: 39021152 |
|---|---|
| 2 |
Qin Y, et al. Front Plant Sci. 2023 Apr 25;14:1164416. doi: 10.3389/fpls.2023.1164416. eCollection 2023. PMID: 37180388 |
| 3 |
Identification of RNA silencing suppressor encoded by citrus chlorotic dwarf-associated virus. Ye X, et al. Front Microbiol. 2024 Jan 25;15:1328289. doi: 10.3389/fmicb.2024.1328289. eCollection 2024. PMID: 38333582 |
| 4 |
Chen Y, et al. Mol Plant Pathol. 2025 Aug;26(8):e70133. doi: 10.1111/mpp.70133. PMID: 40773494 |
| 5 |
Discovery of a novel geminivirus associated with camellia chlorotic dwarf disease. Zhang S, et al. Arch Virol. 2018 Jun;163(6):1709-1712. doi: 10.1007/s00705-018-3780-3. Epub 2018 Mar 2. PMID: 29500570 |
| 6 |
Loconsole G, et al. Virology. 2012 Oct 10;432(1):162-72. doi: 10.1016/j.virol.2012.06.005. Epub 2012 Jul 1. PMID: 22749878 |
| 7 |
Geminiviruses: Taxonomic Structure and Diversity in Genomic Organization. Kulshrestha S, et al. Recent Pat Biotechnol. 2020;14(2):86-98. doi: 10.2174/1872208313666191203100851. PMID: 31793424 |
| 8 |
Updating the Quarantine Status of Prunus Infecting Viruses in Australia. Kinoti WM, et al. Viruses. 2020 Feb 23;12(2):246. doi: 10.3390/v12020246. PMID: 32102210 |
| 9 |
Capulavirus and Grablovirus: two new genera in the family Geminiviridae. Varsani A, et al. Arch Virol. 2017 Jun;162(6):1819-1831. doi: 10.1007/s00705-017-3268-6. Epub 2017 Feb 17. PMID: 28213872 |
| 10 |
Ma Y, et al. J Gen Virol. 2015 Aug;96(8):2421-2434. doi: 10.1099/vir.0.000175. Epub 2015 May 6. PMID: 25953916 |