Tomato severe leaf curl virus
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_000845785.1 |
| Isolate | Guatemala:Sansirisay |
| Release date | 2015/2/12 |
| Submitter | Nakhla,M.K., Maxwell,M.D., Hidayat,S.H., Lange,D.R., Loniello,A.O., Rojas,M.R., Maxwell,D.P., Kitajima,E.W., Rojas,A., Anderson,P., Gilbertson,R.L., Mejia,L., Ramirez,P., Karkashian,J.P., Doyle,M.M. |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
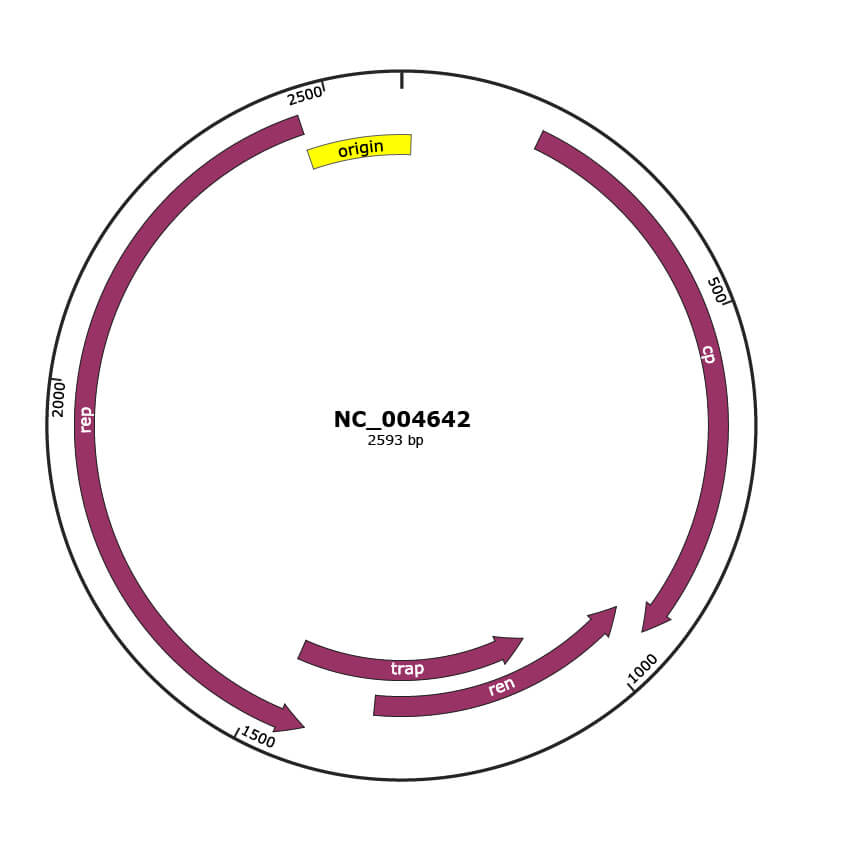
JBrowse
Genome
NC_004642
Gene Information
| NCBI Accession | NP_808780.1 |
|---|---|
| Location | 186-941 |
| Gene Name | cp |
| Protein Name | capsid protein |
| Coding Region | ATGCCTAAGCGTGATGCCCCATGGCGCTTAATGGCGGGTACCCCTAAGGTGAGTCGGTCCTCCAACCAGTCTCCCCGGACTGGTACTGGGCCTAAATTTGATAAGGCCCATGCCTGGGTTAACAGGCCCATGTATAGGAAGCCCAGGATATATCGGGCTTGGAGAACGCCCGACGTTCCAAGGGGCTGTGAAGGCCCATGTAAGGTCCAGTCCTACGAGCAACGACACGATATCTCCCACGTGGGGAAGGTCATGTGCATATCCGATGTCACACGTGGTAATGGTATCACCCACCGTGTTGGTAAGCGGTTTTGTGTTAAGTCTGTGTATATCCTCGGTAAGATATGGATGGACGAGAACATCAAGCTGAAGAACCACACGAACAGTGTCATGTTCTGGTTAGTCAGGGACCGTAGACCCTATGGTACGCCTATGGATTTCGGCCAGGTGTTCAACATGTTTGACAATGAGCCCAGCACTGCCACGGTCAAGACCGATCTACGCGATCGTTACCAGGTCATGCACAAGTTCTATGGCAAGGTGACAGGTGGACAGTATGCCAGCAACGAGCAGGCTATAGTCAAGCGATTCTGGAGGGTCAACAACCATGTGGTCTACAATCACCAAGAGGCTGGCAAGTATGAGAATCACACGGAGAACGCCCTGTTATTGTATATGGCATGTACTCATGCCTCTAACCCTGTGTATGCTACTCTTAAGATCCGGATCTATTTTTATGATTCGGTATTGAATTAA |
| Protein Sequence | MPKRDAPWRLMAGTPKVSRSSNQSPRTGTGPKFDKAHAWVNRPMYRKPRIYRAWRTPDVPRGCEGPCKVQSYEQRHDISHVGKVMCISDVTRGNGITHRVGKRFCVKSVYILGKIWMDENIKLKNHTNSVMFWLVRDRRPYGTPMDFGQVFNMFDNEPSTATVKTDLRDRYQVMHKFYGKVTGGQYASNEQAIVKRFWRVNNHVVYNHQEAGKYENHTENALLLYMACTHASNPVYATLKIRIYFYDSVLN |
| NCBI Accession | NP_808781.1 |
|---|---|
| Location | 938-1336 |
| Gene Name | ren |
| Protein Name | replication enhancer protein |
| Coding Region | ATGGATTCACGCACAGGGGAGAGCATCACTGCGGATCAGGCAGAGAATTCCGTTTTTATCTGGGAGGTACCAAATCCCCTTTATTTCACGATAATCAATGTAGAGGACCCAATTTACAGCAACACCAGGATATACCACATCCAAATACGGTTCAACCACAACCTCAGGAAGGCGCTGAGTCTACACAAAGCCTACCTCAACTACCAAATCTGGACGACATCAGTTCAAGCTTCTGGGACGACTTACTTAAGTAGATTTAAATATTTAGTCTTATTGTATTTAGATAGGTTAGGTGTTATTTGCATTAACAATGTTATCAGAGCTGTACGATTCGCGACAAACAGGTCGTATGTATCTCATGTACTGGAAAATCATTCAATAAAATTCAAATTTTATTAA |
| Protein Sequence | MDSRTGESITADQAENSVFIWEVPNPLYFTIINVEDPIYSNTRIYHIQIRFNHNLRKALSLHKAYLNYQIWTTSVQASGTTYLSRFKYLVLLYLDRLGVICINNVIRAVRFATNRSYVSHVLENHSIKFKFY |
| NCBI Accession | NP_808782.1 |
|---|---|
| Location | 1083-1469 |
| Gene Name | trap |
| Protein Name | transcription activation protein |
| Coding Region | ATGCAAAATTCATCTTCCTCAACTCCACCCTCTATCAAGCCTAAACACAAAATTGCAAAGTCCAGGATTCGTCGAAAGAGAATAGACCTAAACTGTGGCTGCTCGTTCTACCAGCACATCGACTGCGCCAACCATGGATTCACGCACAGGGGAGAGCATCACTGCGGATCAGGCAGAGAATTCCGTTTTTATCTGGGAGGTACCAAATCCCCTTTATTTCACGATAATCAATGTAGAGGACCCAATTTACAGCAACACCAGGATATACCACATCCAAATACGGTTCAACCACAACCTCAGGAAGGCGCTGAGTCTACACAAAGCCTACCTCAACTACCAAATCTGGACGACATCAGTTCAAGCTTCTGGGACGACTTACTTAAGTAG |
| Protein Sequence | MQNSSSSTPPSIKPKHKIAKSRIRRKRIDLNCGCSFYQHIDCANHGFTHRGEHHCGSGREFRFYLGGTKSPLFHDNQCRGPNLQQHQDIPHPNTVQPQPQEGAESTQSLPQLPNLDDISSSFWDDLLK |
| NCBI Accession | NP_808783.1 |
|---|---|
| Location | 1426-2460 |
| Gene Name | rep |
| Protein Name | replication-associated protein Rep |
| Coding Region | ATGCCACGGAACCCTAATTTATTTCGTCTAGCTGCCAAAAATATTTTCTTAACATATCCACAGTGTGATATCCCCAAAGATGAAGCTATTGAAATGCTTCAAAATCTGCCATGGTCAGTCGTCAAACCAACGTACATACGAGTCGCCAGAGAGGAACACGCAGATGGATTTCCGCACCTCCACTGTCTCATCCAACTCTCCGGGAAGTCCAACATCAAGGATGCTAGATTTTTCGACCTCACTCACCCCAGAAGGTCTGCCAATTTTCATCCAAACGCACAGGCAGCCAAAGACACCAACGCCGTCCAGAATTACATCACCAAAGAAGGTGATTATTGTGAATCCGGACAATACAAGGTGTCTGGGGGTACAAAATCAAACAAAGACGACGTCTACCACAACGCCGTCAATGCAGCTAGTGCTGGAGAGGCTCTCGACATTATAAGGGCCGGAGACCCAAAGACGTTCATTGTAAGCTACCATAACGTCAAAGCTAACATCGAGCGGCTCTTTCAAAAGGCTCCGGAACCCTGGGTTCCTCCGTTTCCACTCTCGTCGTTCACGAACGTTCCGGACGAGATGCAAGAATGGGCAGATGATTATTTTGGTAGGGATGCCGCTGCGCGGCCGGAGAGACCTATAAGTATAATCATCGAGGGTGAATCGAGGACGGGAAAGACAATGTGGGCGCGTGCCTTAGGCCCACACAATTATTTGAGTGGACACCTAGATTTCAATTCTAGGGTTTACTCAAATGAAGCGGAATACAACGTCATTGATGACATCACTCCGCAATATTTAAAGATGAAGCATTGGAAAGAGCTGATTGGGGCCCAAAAGGACTGGCAATCAAATTGCAAATACGGGAAGCCAGTTCAAATTAAAGGGGGAATACCGTCAATCGTGCTCTGCAATCCTGGTGAGGGGGCCAGCTATAAAAGTTTCCTCGAAAAAGAGGAAAACAAAGCACTAAAAGATTGGACACTCCACAATGCAAAATTCATCTTCCTCAACTCCACCCTCTATCAAGCCTAA |
| Protein Sequence | MPRNPNLFRLAAKNIFLTYPQCDIPKDEAIEMLQNLPWSVVKPTYIRVAREEHADGFPHLHCLIQLSGKSNIKDARFFDLTHPRRSANFHPNAQAAKDTNAVQNYITKEGDYCESGQYKVSGGTKSNKDDVYHNAVNAASAGEALDIIRAGDPKTFIVSYHNVKANIERLFQKAPEPWVPPFPLSSFTNVPDEMQEWADDYFGRDAAARPERPISIIIEGESRTGKTMWARALGPHNYLSGHLDFNSRVYSNEAEYNVIDDITPQYLKMKHWKELIGAQKDWQSNCKYGKPVQIKGGIPSIVLCNPGEGASYKSFLEKEENKALKDWTLHNAKFIFLNSTLYQA |
References More References in PubMed
| 1 |
Tomato leaf curl New Delhi virus: an emerging plant begomovirus threatening cucurbit production. Cai L, et al. aBIOTECH. 2023 Oct 25;4(3):257-266. doi: 10.1007/s42994-023-00118-4. eCollection 2023 Sep. PMID: 37970471 |
|---|---|
| 2 |
Tomato Yellow Leaf Curl Sardinia Virus Increases Drought Tolerance of Tomato. Sacco Botto C, et al. Int J Mol Sci. 2023 Feb 2;24(3):2893. doi: 10.3390/ijms24032893. PMID: 36769211 |
| 3 |
Mastrochirico M, et al. Plants (Basel). 2023 Jun 21;12(13):2399. doi: 10.3390/plants12132399. PMID: 37446959 |
| 4 |
Zaidi SS, et al. Mol Plant Pathol. 2017 Sep;18(7):901-911. doi: 10.1111/mpp.12481. Epub 2016 Oct 17. PMID: 27553982 |
| 5 |
Host Species-Dependent Transmission of Tomato Leaf Curl New Delhi Virus-ES by Bemisia tabaci. Janssen D, et al. Plants (Basel). 2022 Jan 30;11(3):390. doi: 10.3390/plants11030390. PMID: 35161372 |
| 6 |
Naganur P, et al. Plants (Basel). 2023 Jan 20;12(3):490. doi: 10.3390/plants12030490. PMID: 36771575 |
| 7 |
Bottle gourd IC-0262269, a super-susceptible genotype to tomato leaf curl Palampur virus. Nayaka SN, et al. 3 Biotech. 2024 Jan;14(1):8. doi: 10.1007/s13205-023-03838-y. Epub 2023 Dec 8. PMID: 38074288 |
| 8 |
Vo TTB, et al. Microorganisms. 2023 Dec 1;11(12):2907. doi: 10.3390/microorganisms11122907. PMID: 38138051 |
| 9 |
Dufulin Impacts Plant Defense Against Tomato Yellow Leaf Curl Virus Infecting Tomato. Huang L, et al. Viruses. 2024 Dec 31;17(1):53. doi: 10.3390/v17010053. PMID: 39861842 |
| 10 |
Coat protein is responsible for tomato leaf curl New Delhi virus pathogenicity in tomato. Vo TTB, et al. Front Plant Sci. 2023 Jul 10;14:1206255. doi: 10.3389/fpls.2023.1206255. eCollection 2023. PMID: 37492775 |