Tomato mild mosaic virus
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_000874565.1 |
| Isolate | Brazil |
| Release date | 2015/2/13 |
| Submitter | Castillo-Urquiza,G.P., Beserra,J.E. Jr., Bruckner,F.P., Lima,A.T., Varsani,A., Alfenas-Zerbini,P., Murilo Zerbini,F., Beserra,J.E.A. Jr., Lima,A.T.M., Zerbini,P.A., Zerbini,F.M. |
| Host | |
| Download | Genome |GFF3 |PEP |CDS |
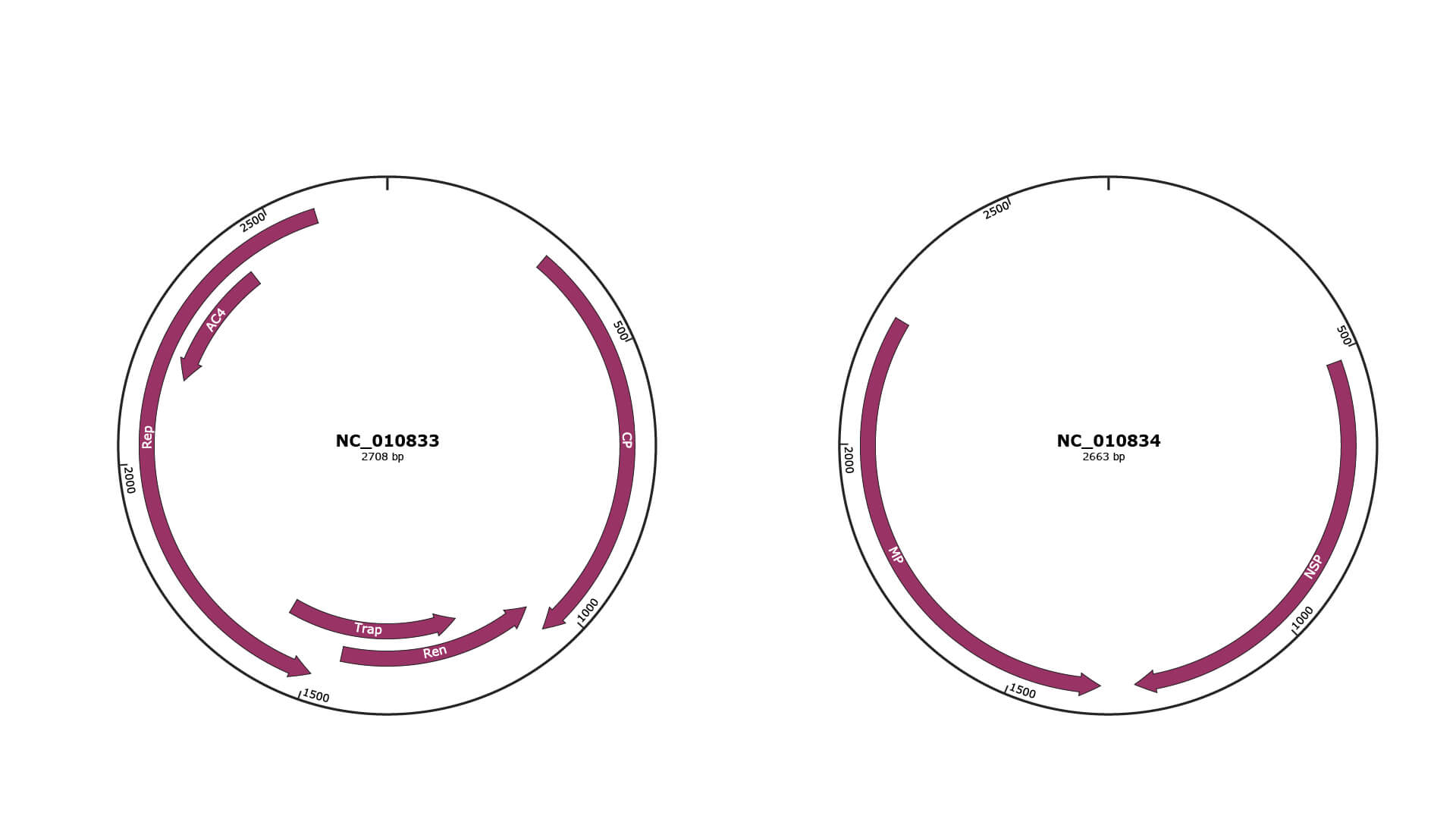
Genomic Organization
JBrowse
Genome
NC_010833
NC_010834
Gene Information
| NCBI Accession | YP_001960945.1 |
|---|---|
| Location | 302-1051 |
| Gene Name | CP |
| Protein Name | coat protein |
| Coding Region | ATGCCGAAGCGGGACGCCCCATGGCGCCTGATGGCGGGAACCTCAAAGGTTAGCCGATCTGTCAACTACTCCCCTCGTGGAGGCCCAAAGTTCAACAAGGCATCTGAGTGGGTCAACAGGCCCATGTATAGAAAGCCCAGGATATATCGGACGCTAAGGTCTCCTGACGTCCCCAAAGGCTGTGAAGGGCCTTGTAAGGTCCAGTCGTTCGAGGCGCGACATGATGTCTCTCATGTCGGGAAGGTCATATGCGTCTCTGACGTGACACGTGGTAATGGTATTACCCATCGTGTGGGTAAGCGTTTCTGTGTTAAGTCTGTATATATTTTAGGGAAGATCTGGATGGACGAAGCGATCAAGCTCAAGAACCACACGAACAGCGTCATGTTCTGGTTAGTTAGAGACAGGAGACCCTACGGAACTCCTATGGATTTTGGACAGGTGTTCAACCTGTTCGACAACGAGCCCAGCACTGCCACGGTTAAGAGCGATCTCCGTGATCGCTTCCAAGTGATGCGCAAGTTTTACGCCAAGGTCACAGGTGGACAGTATGCTAGCAACGAGCAGGCTCTGGTCAAGCGTTTCTGGAAGGTCAACAATCATGTGGTCTACAACCACCAAGAAGCTGCGAAGTACGAGAACCACACAGAGAACGCTATGTTATTGTATATGGCATGTACCCATGCCTCTAACCCTGTGTATGCGAGTCTCAAGATTCGAATCTACTTCTACGATTCCGTTCTCAATTAA |
| Protein Sequence | MPKRDAPWRLMAGTSKVSRSVNYSPRGGPKFNKASEWVNRPMYRKPRIYRTLRSPDVPKGCEGPCKVQSFEARHDVSHVGKVICVSDVTRGNGITHRVGKRFCVKSVYILGKIWMDEAIKLKNHTNSVMFWLVRDRRPYGTPMDFGQVFNLFDNEPSTATVKSDLRDRFQVMRKFYAKVTGGQYASNEQALVKRFWKVNNHVVYNHQEAAKYENHTENAMLLYMACTHASNPVYASLKIRIYFYDSVLN |
| NCBI Accession | YP_001960946.1 |
|---|---|
| Location | 1048-1446 |
| Gene Name | Ren |
| Protein Name | replication enhancer protein |
| Coding Region | ATGGATTCACGCACCGGGGAGAGCATCACTGCACCTCAGGCAGAGAATGGCGTATATATCTGGGAGATATCAAATCCCCTCTATTTCAAGATATACAACGTAGAGGATCTACGGTACACGGCGACCAGAGTGTACCACATCCAGATACGGTTCAACCACAACCTCAGGAGGATGCTGGGTCTACACAAGGCCTTCCTGAACTTCCAAGTCTGGACGACATCAGTTCGAGCTTCTGGGACGACATATTTACATAGGTTTAGGCAGTTAGTTATGTTGTATCTCGATCAATTAGGTGTGATCTCCCTTAATAATGTAATTAGGGCTGTTAGGTTTGCAACAGACAAACCATATGTAAACTATGTACTGGAAAATCATTCAATAAAATTTAAATTTTATTAA |
| Protein Sequence | MDSRTGESITAPQAENGVYIWEISNPLYFKIYNVEDLRYTATRVYHIQIRFNHNLRRMLGLHKAFLNFQVWTTSVRASGTTYLHRFRQLVMLYLDQLGVISLNNVIRAVRFATDKPYVNYVLENHSIKFKFY |
| NCBI Accession | YP_001960947.1 |
|---|---|
| Location | 1193-1582 |
| Gene Name | Trap |
| Protein Name | trans-activating protein |
| Coding Region | ATGCTAAATTCATCTTCCTCGACGCCCCCCTCTATCAAGCCCAAGCACAGGATTGCGAAGAAGAGGGCTACCAGACGACGCAGAATTGACCTAGACTGCGGGTGCTCGGTCTTCGTGCACCTTAACTGCGCAGGGCATGGATTCACGCACCGGGGAGAGCATCACTGCACCTCAGGCAGAGAATGGCGTATATATCTGGGAGATATCAAATCCCCTCTATTTCAAGATATACAACGTAGAGGATCTACGGTACACGGCGACCAGAGTGTACCACATCCAGATACGGTTCAACCACAACCTCAGGAGGATGCTGGGTCTACACAAGGCCTTCCTGAACTTCCAAGTCTGGACGACATCAGTTCGAGCTTCTGGGACGACATATTTACATAG |
| Protein Sequence | MLNSSSSTPPSIKPKHRIAKKRATRRRRIDLDCGCSVFVHLNCAGHGFTHRGEHHCTSGREWRIYLGDIKSPLFQDIQRRGSTVHGDQSVPHPDTVQPQPQEDAGSTQGLPELPSLDDISSSFWDDIFT |
| NCBI Accession | YP_001960948.1 |
|---|---|
| Location | 1494-2579 |
| Gene Name | Rep |
| Protein Name | replication-associated protein |
| Coding Region | ATGCCACCGCCCAAACGTTTTAGAATAAATGCCAAGAACTATTTCTTGACATACCCCAAATGCAGCCTAACCAAAGAAGAGGCACTTTCCCAATTATTAGCCCTAGAAATTCCTGCACAGAAGAAATTTATCAAGGTAGCTAGAGAGCTCCACGACGATGGGGAGCCTCATCTCCACGTCCTCATCCAGTTCGAAGGGAAATTCCAGTGCACGAATAATAGATTCTTCGACCTTGTCTCCCCAACCAGGTCAGCACATTTCCATCCGAACGTTCAGGGGGCTAAGTCGTCGTCCGACGTCAAGTCCTATGTCGACAAGGACGGAGATACAGTCGAATGGGGAGTTTTCCAGGTCGACGGAAGAAGTGCTAGAGGAGGTTGCCAGACAGCTAACGACGCTGCCGCAGAGGCCTTAAACGCGCCGTCGAAAGAAGCGGCGCTGAGCATAATCAGAGAGAAGTTGCCGGAAAAGTATCTCTTCCAATTCCATAATCTAAATAGTAATTTAGATCGCATTTTCGCAAAGGCTCCGGAAACTTGGTCTCCTCCGTTTCCACTCTCCTCATTCACTCACGTTCCCGACGAGATGCAAGAGTGGGCCGATGACTATTTTGGAAGAGATGCCGCTGCGCGGCCCAGCAGGCCTGTTAGTTTGATAGTCGAAGGAGATAGTCGAACAGGCAAGACAATGTGGGCTCGTGCTTTAGGGGCCCATAACTATCTCAGTGGACACCTGGACTTCAACTCCAGGGTCTATTCAAATGAGGCTGAATATAACGTCATTGATGACGTCCCTCCGCACTACCTAAAGCTAAAGCACTGGAAAGAATTGATGGGAGCTCAAAAGGACTGGCAGTCAAATTGCAAATACGGAAAGCCTGTTCAAATTAAAGGCGGTATCCCATCAATCGTGCTCTGCAATCCTGGAGAGGGGGCCAGCTATAAAGATTTCCTCGAGAAAGAGGAAAACCTTGCTCTGAAGAACTGGACGATCAAGAATGCTAAATTCATCTTCCTCGACGCCCCCCTCTATCAAGCCCAAGCACAGGATTGCGAAGAAGAGGGCTACCAGACGACGCAGAATTGA |
| Protein Sequence | MPPPKRFRINAKNYFLTYPKCSLTKEEALSQLLALEIPAQKKFIKVARELHDDGEPHLHVLIQFEGKFQCTNNRFFDLVSPTRSAHFHPNVQGAKSSSDVKSYVDKDGDTVEWGVFQVDGRSARGGCQTANDAAAEALNAPSKEAALSIIREKLPEKYLFQFHNLNSNLDRIFAKAPETWSPPFPLSSFTHVPDEMQEWADDYFGRDAAARPSRPVSLIVEGDSRTGKTMWARALGAHNYLSGHLDFNSRVYSNEAEYNVIDDVPPHYLKLKHWKELMGAQKDWQSNCKYGKPVQIKGGIPSIVLCNPGEGASYKDFLEKEENLALKNWTIKNAKFIFLDAPLYQAQAQDCEEEGYQTTQN |
| NCBI Accession | YP_001960949.1 |
|---|---|
| Location | 2165-2422 |
| Gene Name | AC4 |
| Protein Name | AC4 |
| Coding Region | ATGGGGAGCCTCATCTCCACGTCCTCATCCAGTTCGAAGGGAAATTCCAGTGCACGAATAATAGATTCTTCGACCTTGTCTCCCCAACCAGGTCAGCACATTTCCATCCGAACGTTCAGGGGGCTAAGTCGTCGTCCGACGTCAAGTCCTATGTCGACAAGGACGGAGATACAGTCGAATGGGGAGTTTTCCAGGTCGACGGAAGAAGTGCTAGAGGAGGTTGCCAGACAGCTAACGACGCTGCCGCAGAGGCCTTAA |
| Protein Sequence | MGSLISTSSSSSKGNSSARIIDSSTLSPQPGQHISIRTFRGLSRRPTSSPMSTRTEIQSNGEFSRSTEEVLEEVARQLTTLPQRP |
| NCBI Accession | YP_001960950.1 |
|---|---|
| Location | 518-1285 |
| Gene Name | NSP |
| Protein Name | nuclear shuttle protein |
| Coding Region | ATGTATCCTGTTAAGTATAGACGTGGCTGGTCGACTACTCAACGACGAAGTTATCGACGGGCTCCTGTGTTCAAGCGGAATGCCGTTAAACGTGCAGATTGGATACGTCGACCGAGTAATTCAATGAAGGCCCATGACGAGCCCAAGATGACAGCCCAGCGGATACATGAGAACCAGTTTGGCCCAGAATTCATGATGGTTCAGAATACAGCAATTTCTACTTACATATCCTTTCCCAATCTTGGTAAGACAGAACCGAACCGGTCTAGGTCATATATTAAGCTAAAACGACTTCGTTTTAAAGGGACTGTCAAGATAGAACGTGTTCAGCCAGATGTCAACATGGACGGTTCGGTCTCGAAAACCGAAGGTGTGTTCTCTCTCGTAGTGGTTGTGGATCGTAAGCCTCATTTGGGCCCTTCTGGATGTTTGCACACCTTTGACGAGCTCTTCGGTGCAAGGATCCACAGCCATGGGAACCTATCTATTACTCCTTCATTGAAGGACCGGTACTACATACGTCATGTGACCAAACATGTACTCTCGGCGGAAAAGGACACCATGATGGTCAACCTCGAAGGGACGACATTCCTATCTAACAGGCGTGTTAGCTGTTGGGCCGGTTTTAAGGATCATGATCATGATTCATGTAATGGGGTTTATGCTAACATAAGCAAGAACGCCCTGTTAGTTTATTACTGTTGGATGTCGGATATTATGTCCAAGGCATCGACATTTGTATCATATGATCTGGATTATGTTGGTTGA |
| Protein Sequence | MYPVKYRRGWSTTQRRSYRRAPVFKRNAVKRADWIRRPSNSMKAHDEPKMTAQRIHENQFGPEFMMVQNTAISTYISFPNLGKTEPNRSRSYIKLKRLRFKGTVKIERVQPDVNMDGSVSKTEGVFSLVVVVDRKPHLGPSGCLHTFDELFGARIHSHGNLSITPSLKDRYYIRHVTKHVLSAEKDTMMVNLEGTTFLSNRRVSCWAGFKDHDHDSCNGVYANISKNALLVYYCWMSDIMSKASTFVSYDLDYVG |
| NCBI Accession | YP_001960951.1 |
|---|---|
| Location | 1346-2227 |
| Gene Name | MP |
| Protein Name | movement protein |
| Coding Region | ATGGAATCTCAGTTAGCTAATCCTCCGAACGCCTTCAATTATATAGAGTCGCAAAGGGACGAGTACCAGCTGTCTCATGACCTAACTGAGATCGTACTGCAGTTTCCGTCCACCGCCTCCCAGATAAGTGCCAGACTCAGTCGTAGATGTATGAAAATAGACCACTGCGTCATAGAGTACAGGCAGCAGGTTCCGATTAACGCCACTGGGGCGGTGGTAGTGGAGATTCACGACAAGAGGATGACGGACAATGAGTCGTTACAGGCGTCGTGGACTTTCCCGATCAGGTGTAACATCGATCTCCACTACTTTTCATCTTCCTTCTTCTCCCTCAAAGACCCAATTCCATGGAAGCTGTACTACAGAGTTTGCGACACGAACGTCCATCAGAGGACCCACTTCGCAAAATTCAAAGGTAAACTGAAGCTGTCGACGGCTAAACACTCCGTGGATATTCCTTTCAGGGCTCCGACGGTCAAGATCCTGTCCAAACAGTTTACCGACAAGGATGTCGATTTCTGCCACGTGGGATACGGAAGATGGGAGAGGAAACCAGTCCGATCCGCATCCGCCTCAACAATTGGGCTTCGCAGCCCAATACAACTTAGGCCAGGAGAGTCGTGGGCTGTCAGGAGCACTGTAGGTGCCAATCCATCGGATGCGGAGTCCGATATAGTAGAGACATCGCATCCATACAGGGAGCTGAACAGACTGGGTACAACAATGTTGGACCCAGGCGAATCGGCTTCCATCGTGGGGGCCCAGAGAGCCCAGTCCAACATTACGATGTCTCTGGGACAATTAAACGAACTGCTGAGGAACACGGTCCAGGAGTGCATTAACAGTAACTGTGTGCCTTCGCAGGCCAAGTCTTTAAATTAA |
| Protein Sequence | MESQLANPPNAFNYIESQRDEYQLSHDLTEIVLQFPSTASQISARLSRRCMKIDHCVIEYRQQVPINATGAVVVEIHDKRMTDNESLQASWTFPIRCNIDLHYFSSSFFSLKDPIPWKLYYRVCDTNVHQRTHFAKFKGKLKLSTAKHSVDIPFRAPTVKILSKQFTDKDVDFCHVGYGRWERKPVRSASASTIGLRSPIQLRPGESWAVRSTVGANPSDAESDIVETSHPYRELNRLGTTMLDPGESASIVGAQRAQSNITMSLGQLNELLRNTVQECINSNCVPSQAKSLN |
References More References in PubMed
| 1 |
Engineered Resistance to Tobamoviruses. Carr JP. Viruses. 2024 Jun 22;16(7):1007. doi: 10.3390/v16071007. PMID: 39066170 |
|---|---|
| 2 |
Zhang S, et al. Mol Plant Pathol. 2022 Sep;23(9):1262-1277. doi: 10.1111/mpp.13229. Epub 2022 May 22. PMID: 35598295 |
| 3 |
Tomato Brown Rugose Fruit Virus Contributes to Enhanced Pepino Mosaic Virus Titers in Tomato Plants. Klap C, et al. Viruses. 2020 Aug 11;12(8):879. doi: 10.3390/v12080879. PMID: 32796777 |
| 4 |
Nowakowska M, et al. Int J Mol Sci. 2025 Dec 4;26(23):11749. doi: 10.3390/ijms262311749. PMID: 41373893 |
| 5 |
Giakountis A, et al. Plant Dis. 2018 Nov;102(11):2268-2276. doi: 10.1094/PDIS-08-17-1259-RE. Epub 2018 Sep 6. PMID: 30189158 |
| 6 |
Hanssen IM, et al. Plant Physiol. 2011 May;156(1):301-18. doi: 10.1104/pp.111.173906. Epub 2011 Mar 22. PMID: 21427280 |
| 7 |
European Food Safety Authority (EFSA). EFSA J. 2017 Jan 23;15(1):e04651. doi: 10.2903/j.efsa.2017.4651. eCollection 2017 Jan. PMID: 32625250 |
| 8 |
Elbeshehy EKF, et al. Molecules. 2022 Dec 24;28(1):139. doi: 10.3390/molecules28010139. PMID: 36615333 |
| 9 |
A New Technique to Select Mild Strains of Cucumber mosaic virus. Kobori T, et al. Plant Dis. 2005 Aug;89(8):879-882. doi: 10.1094/PD-89-0879. PMID: 30786521 |
| 10 |
Zhao W, et al. J Virol Methods. 2021 Dec;298:114277. doi: 10.1016/j.jviromet.2021.114277. Epub 2021 Sep 4. PMID: 34492235 |