Tomato leaf curl Tanzania virus
Basic Information
| Genus | Begomovirus |
|---|---|
| Isolate | TZ-Ten-05 |
| Vector | |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
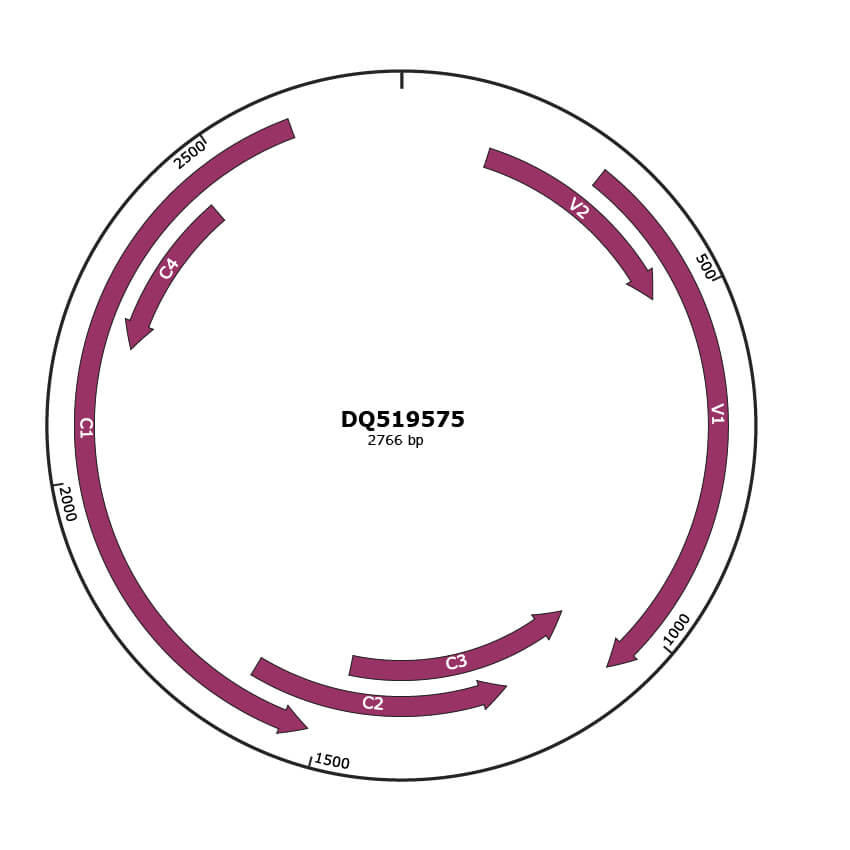
JBrowse
Genome
DQ519575
Gene Information
| NCBI Accession | ABF61898.1 |
|---|---|
| Location | 137-487 |
| Gene Name | V2 |
| Protein Name | pre-coat protein |
| Coding Region | ATGTGGGATCCGTTAGTAAATGAGTTTCCAGACTCTGTTCACGGGTTTCGTTGTATGCTTGCAATAAAATATTTACAGGCCGTTGAGTCCACTTACGAGCCCAATACATTGGGCCACGATTTAATTCGAGATCTCATTTGTGTCGTTAGAGCCAGAGATTATGTCGAAGCGACCCGGAGATATAATCATTTCAACGCCCGCCTCGAAGGTGCGTCGAAGGCTGAACTTCGACAACCCGTATACCAGCCGTGCTGCTGTCCCCATCGTCCCAGGCACAAGCAGACGTCGATCATGGACTTACAGGCCCATGTATCGAAAGCCCAGGATGTACAGAATGTACAGAAGCCCTGA |
| Protein Sequence | MWDPLVNEFPDSVHGFRCMLAIKYLQAVESTYEPNTLGHDLIRDLICVVRARDYVEATRRYNHFNARLEGASKAELRQPVYQPCCCPHRPRHKQTSIMDLQAHVSKAQDVQNVQKP |
| NCBI Accession | ABF61895.1 |
|---|---|
| Location | 297-1073 |
| Gene Name | V1 |
| Protein Name | coat protein |
| Coding Region | ATGTCGAAGCGACCCGGAGATATAATCATTTCAACGCCCGCCTCGAAGGTGCGTCGAAGGCTGAACTTCGACAACCCGTATACCAGCCGTGCTGCTGTCCCCATCGTCCCAGGCACAAGCAGACGTCGATCATGGACTTACAGGCCCATGTATCGAAAGCCCAGGATGTACAGAATGTACAGAAGCCCTGATGTTCCTCGGGGTTGTGAAGGCCCATGTAAGGTTCAGTCTTATGAGCAGAGGGATGATGTAAAGCATACTGGTATTGTTCGTTGTGTTAGTGATGTAACTAGAGGTAATGGAATTACTCATAGAGTAGGAAAACGGTTCTGCATTAAGTCCATATATATTTTAGGAAAAATATGGATGGATGAGAATATCAAGAAGCAGAACCATACTAATCAGGTCATGTTTTTTTTGGTCCGTGATAGAAGGCCCTATGGCCCAAGCCCAATGGATTTTGGGCAGGTGTTTAACATGTTTGATAATGAGCCCAGTACAGCCACTGTGAAGAATGACCTCCGAGACAGATATCAAGTTTTGCGGAAATTTCATGCAACTGTTGTCGGTGGACCATCTGGGATGAAAGAGCAGGCATTACTTAAAAGATTTTTTAGAATTAATAATCATGTAGTTTATAATCATCAAGAGAATGCTAAGTATGAGAATCATACTGAGAATGCTCTGTTGTTGTATATGGCATGTACTCATGCTTCTAACCCTGTGTATGCTACGTTGAAAATACGTATCTATTTCTATGTTTCAGTTGGGAATTAA |
| Protein Sequence | MSKRPGDIIISTPASKVRRRLNFDNPYTSRAAVPIVPGTSRRRSWTYRPMYRKPRMYRMYRSPDVPRGCEGPCKVQSYEQRDDVKHTGIVRCVSDVTRGNGITHRVGKRFCIKSIYILGKIWMDENIKKQNHTNQVMFFLVRDRRPYGPSPMDFGQVFNMFDNEPSTATVKNDLRDRYQVLRKFHATVVGGPSGMKEQALLKRFFRINNHVVYNHQENAKYENHTENALLLYMACTHASNPVYATLKIRIYFYVSVGN |
| NCBI Accession | ABF61897.1 |
|---|---|
| Location | 1070-1474 |
| Gene Name | C3 |
| Protein Name | replication enhancement protein |
| Coding Region | ATGGATTTACGCACCGGGGAACTCATCACTGCGCATCAGGCACAGAGTGGCGTCTATACCTGGGAAATCGCCAATCCCCTATATTTCAGCATCACGGAACACTCGAAGAGACCATTCAACTACAACCACGACATCATCACAGTACAACTCCGATTCAACCACAACCTAAGGAAGGAACTGGGGATTCACAAATGTTTCATGACCTTCAAGATCTGGACAACCTTACAGCCTCAGACTGGTCTTTTCTTAAGAGTCTTTAAGACCCAAGTTAGAAAATATTTAAACAATCTTGGTGTAATTTCAATTAATAATGTAATTAGAGCAATAGAATATGTATTGTTTGATGTAATCAACGGGACCTTATGGGTGGAACAAAATTATGATATAAAATTCAAACTTTATTAA |
| Protein Sequence | MDLRTGELITAHQAQSGVYTWEIANPLYFSITEHSKRPFNYNHDIITVQLRFNHNLRKELGIHKCFMTFKIWTTLQPQTGLFLRVFKTQVRKYLNNLGVISINNVIRAIEYVLFDVINGTLWVEQNYDIKFKLY |
| NCBI Accession | ABF61896.1 |
|---|---|
| Location | 1215-1622 |
| Gene Name | C2 |
| Protein Name | transcriptional activation protein |
| Coding Region | ATGCGATCTTCGTCACCATCCAACAGCCATTGTACTCAGATTCCCATCAAGGTCCAACACAAAACAGCCAAGCAGAGGCTTCAACGGAGGAAGAGGATAGACCTTAATTGCGGGTGCTCGTACTACTTGTCACTCAACTGTCGCAACCATGGATTTACGCACCGGGGAACTCATCACTGCGCATCAGGCACAGAGTGGCGTCTATACCTGGGAAATCGCCAATCCCCTATATTTCAGCATCACGGAACACTCGAAGAGACCATTCAACTACAACCACGACATCATCACAGTACAACTCCGATTCAACCACAACCTAAGGAAGGAACTGGGGATTCACAAATGTTTCATGACCTTCAAGATCTGGACAACCTTACAGCCTCAGACTGGTCTTTTCTTAAGAGTCTTTAA |
| Protein Sequence | MRSSSPSNSHCTQIPIKVQHKTAKQRLQRRKRIDLNCGCSYYLSLNCRNHGFTHRGTHHCASGTEWRLYLGNRQSPIFQHHGTLEETIQLQPRHHHSTTPIQPQPKEGTGDSQMFHDLQDLDNLTASDWSFLKSL |
| NCBI Accession | ABF61894.1 |
|---|---|
| Location | 1516-2610 |
| Gene Name | C1 |
| Protein Name | replication-associated protein |
| Coding Region | ATGCCTAGAGCAGGTCGTTTTAGCATCAAAGCCAAAAATTATTTCCTCACATACCCTAAATGTTCTCTTTCAAAAGAAGAAGCATTAACTCAACTACGAGCCCTTCAAACACCTACAAATAAGCGTTTCATCAAAATTTGCAGAGAGCTTCACGAGAATGGGGAACCTCATCTGCATGCCCTCATTCAGTTCGAAGGAAAGTACAATTGTACCAATAACAGATTCTTCGACCTGGTATCCCCAACCAGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAATCAAGCTCCGACGTCAAGTCCTACATCCACAAGGATGGAGATACACTCGAATGGGGGGAATTCCAGATCGACGGCAGAAGTGCTAGAGGAGGCTGCCATAATGCTAACGACGCATGCGCCGAGGCATTAAACGCAGGATCCGCTGAAGCCGCTTTGGCTATTATTAGGGAGAAACTCCCTAAAGATTTTATTTTTCAGTATCATAATTTAAAGTGTAATTTAGATAGGATTTTTACACCTCCGTTGGAGGATTATGTTTCTCCTTTTTTATGTTCTTCTTTCGATCAAGTTCCAGAAGAGCTTGAAGAGTGGGCGGCGGCGAATGTTGTCAGTACCGCTGCGCGGCCATTGAGACCCATGAGTATTGTGATTGAAGGGGATAGTCGTACCGGGAAAACAATGTGGGCTAGGTCATTAGGACCACACAATTATTTGTGTGGTCATCTTGATCTCAGTCCAAAGGTGTACAGTAATAATGCATGGTACAACGTCATTGATGACGTCGATCCGCATTATCTAAAACACTTCAAGGAATTCATGGGCGCACAAAGGGACTGGCAAAGCAACACCAAGTACGGGAAACCAATTCAAATTAAAGGCGGAATTCCCACTATCTTCCTCTGCAATCCAGGACCAACTTCATCATATAAAGAGTACTTGGACGAGGAAAAGAACGCACCACTAAAGGCGTGGGCAATTAAAAATGCGATCTTCGTCACCATCCAACAGCCATTGTACTCAGATTCCCATCAAGGTCCAACACAAAACAGCCAAGCAGAGGCTTCAACGGAGGAAGAGGATAGACCTTAA |
| Protein Sequence | MPRAGRFSIKAKNYFLTYPKCSLSKEEALTQLRALQTPTNKRFIKICRELHENGEPHLHALIQFEGKYNCTNNRFFDLVSPTRSAHFHPNIQGAKSSSDVKSYIHKDGDTLEWGEFQIDGRSARGGCHNANDACAEALNAGSAEAALAIIREKLPKDFIFQYHNLKCNLDRIFTPPLEDYVSPFLCSSFDQVPEELEEWAAANVVSTAARPLRPMSIVIEGDSRTGKTMWARSLGPHNYLCGHLDLSPKVYSNNAWYNVIDDVDPHYLKHFKEFMGAQRDWQSNTKYGKPIQIKGGIPTIFLCNPGPTSSYKEYLDEEKNAPLKAWAIKNAIFVTIQQPLYSDSHQGPTQNSQAEASTEEEDRP |
| NCBI Accession | ABF61899.1 |
|---|---|
| Location | 2196-2453 |
| Gene Name | C4 |
| Protein Name | C4 protein |
| Coding Region | ATGGGGAACCTCATCTGCATGCCCTCATTCAGTTCGAAGGAAAGTACAATTGTACCAATAACAGATTCTTCGACCTGGTATCCCCAACCAGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAATCAAGCTCCGACGTCAAGTCCTACATCCACAAGGATGGAGATACACTCGAATGGGGGGAATTCCAGATCGACGGCAGAAGTGCTAGAGGAGGCTGCCATAATGCTAACGACGCATGCGCCGAGGCATTAA |
| Protein Sequence | MGNLICMPSFSSKESTIVPITDSSTWYPQPGQHISIRTFRELNQAPTSSPTSTRMEIHSNGGNSRSTAEVLEEAAIMLTTHAPRH |
References More References in PubMed
| 1 |
Avedi EK, et al. Virol J. 2021 Jan 6;18(1):2. doi: 10.1186/s12985-020-01466-z. PMID: 33407584 |
|---|---|
| 2 |
A New Geminivirus Associated with a Leaf Curl Disease of Tomato in Tanzania. Chiang BT, et al. Plant Dis. 1997 Jan;81(1):111. doi: 10.1094/PDIS.1997.81.1.111B. PMID: 30870927 |
| 3 |
Shih SL, et al. Plant Dis. 2006 Dec;90(12):1550. doi: 10.1094/PD-90-1550C. PMID: 30780977 |
| 4 |
Mwaipopo B, et al. Plant Dis. 2018 Nov;102(11):2361-2370. doi: 10.1094/PDIS-01-18-0198-RE. Epub 2018 Sep 19. PMID: 30252625 |
| 5 |
Lanai: A small, fast growing tomato variety is an excellent model system for studying geminiviruses. Rajabu CA, et al. J Virol Methods. 2018 Jun;256:89-99. doi: 10.1016/j.jviromet.2018.03.002. Epub 2018 Mar 9. PMID: 29530481 |
| 6 |
Kyallo M, et al. Arch Virol. 2017 May;162(5):1393-1396. doi: 10.1007/s00705-016-3217-9. Epub 2017 Jan 9. PMID: 28070648 |
| 7 |
Management of whitefly-transmitted viruses in open-field production systems. Lapidot M, et al. Adv Virus Res. 2014;90:147-206. doi: 10.1016/B978-0-12-801246-8.00003-2. PMID: 25410102 |
| 8 |
First Molecular Identification of a Begomovirus Isolated from Tomato in Madagascar. Delatte H, et al. Plant Dis. 2002 Dec;86(12):1404. doi: 10.1094/PDIS.2002.86.12.1404C. PMID: 30818457 |
| 9 |
Zhou X, et al. J Gen Virol. 1998 Nov;79 ( Pt 11):2835-40. doi: 10.1099/0022-1317-79-11-2835. PMID: 9820161 |
| 10 |
Reyes MI, et al. Plant J. 2017 Dec;92(5):796-807. doi: 10.1111/tpj.13716. Epub 2017 Oct 24. PMID: 28901681 |