Tomato leaf curl Sinaloa virus
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_000871805.1 |
| Isolate | Nicaragua:Santa Lucia |
| Release date | 2015/2/13 |
| Submitter | Rojas,A., Kvarnheden,A., Marcenaro,D., Valkonen,J.P. |
| Host | |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
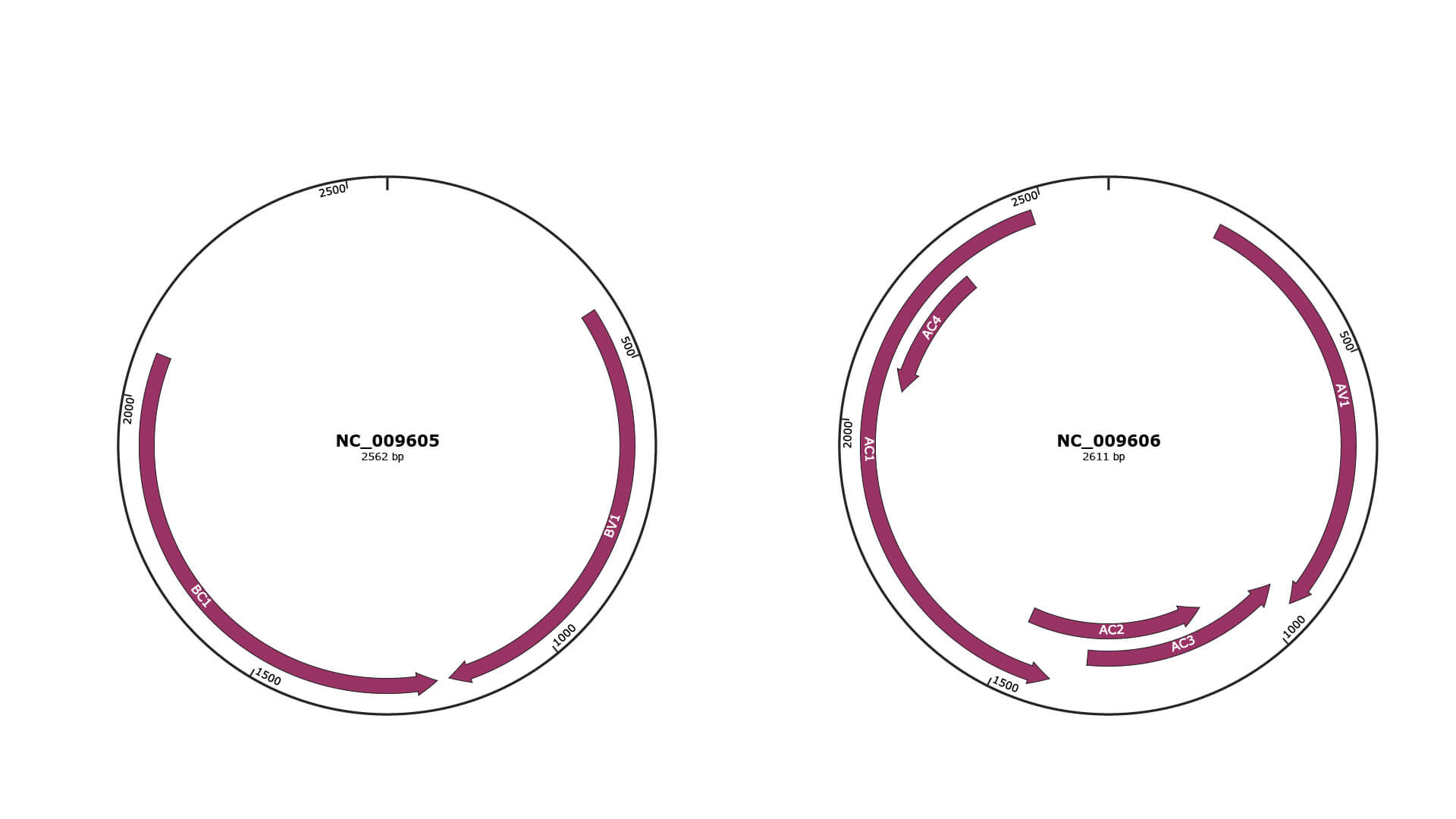
JBrowse
Genome
NC_009605
NC_009606
Gene Information
| NCBI Accession | YP_001294912.1 |
|---|---|
| Location | 405-1175 |
| Gene Name | BV1 |
| Protein Name | nuclear shuttle protein |
| Coding Region | ATGTATTATTTTAGGAATAAACGTGGTTCATCCTTTATTCAACGGCGGTATCATTCACGTAATAATGTGTTTAAGCGTAATACTTCGATCAAACGAGATGATGGTAAACGTCGTCATGTCTATTCTAACAAGTCCAATGATGAGCCAAAGATGTCATCGCAACGTATTCACGAGAACCAGTTTGGTCAAGAGTTTGTGATGGTCCATAATTCTGCCATATCTACGTTTGTAAGCTATCCCAGTCTCGGTAAGACTGAACCGAATCGTAGCAGGTCCTACATTAAGTTGAAACGACTGCGTTTTAAGGGTACTGTGAAGATTGAACGTGTTGTGTCGGAAATGAACATGGACGGTTCTACTTCGAAGGTAGAAGGAGTGTTTTCACTTGTCATCGTAGTGGACCGTAAACCCCACTTGGGTGCATCTGGTTGTCTACATACCTTTGACGAACTCTTTGGAGCTAGGATTCACAGCCATGGTAATCTTAGCATAACACCTTCTCTCAAGGATCGCTTCTATATAAGACATGTGTTCAAACGTGTTCTATCTGTGGAGAAGGATACGACGATGGTGGATGTGGAAGGATCTACCACGCTTTCTAACAGGCGATATAGTTGTTGGTCTACATTTAAGGATTTGGATCACGACTCGTGCAATGGTGTTTATGGAAACATTAGCAAGAACGCTCTCTTAGTTTACTACTGTTGGATGTCAGATACTATGTCTAAGGCATCTACTTTTGTATCGTTTGACCTTGATTATGTCGGATGA |
| Protein Sequence | MYYFRNKRGSSFIQRRYHSRNNVFKRNTSIKRDDGKRRHVYSNKSNDEPKMSSQRIHENQFGQEFVMVHNSAISTFVSYPSLGKTEPNRSRSYIKLKRLRFKGTVKIERVVSEMNMDGSTSKVEGVFSLVIVVDRKPHLGASGCLHTFDELFGARIHSHGNLSITPSLKDRFYIRHVFKRVLSVEKDTTMVDVEGSTTLSNRRYSCWSTFKDLDHDSCNGVYGNISKNALLVYYCWMSDTMSKASTFVSFDLDYVG |
| NCBI Accession | YP_001294913.1 |
|---|---|
| Location | 1196-2077 |
| Gene Name | BC1 |
| Protein Name | movement protein |
| Coding Region | ATGGAATCTCAATTAGCATTTCCTCCTAATGCCTTCAATTATATAGAGTCTCATCGGGACGAATATCAGCTTTCCCATGATTTAACTGAGATAATTCTGCAATTTCCGTCGACGGCGTCACAGTTGACGGCTAGACTAAGTCGGAGCTGCATGAAAATAGACCACTGCGTCATAGAATACAGACAACAAGTCCCGATTAACGCAACTGGGTCGGTGGTAGTGGAGATCCACGACAAAAGAATGACAGACAACGAATCTCTACAGGCATCATGGACTTTTCCGATAAGATGCAACATAGATCTCCACTACTTTTCCGCTTCGTTCTTCTCGTTGAAAGACCCAATTCCATGGAAACTCTATTACAGAGTTTGCGATACAAATGTTCATCAAAGGACCCACTTCGCGAAATTCAAAGGGAAGCTGAAGCTGTCGACGGCGAAACACTCGGTGGACATCCCTTTCCGGGCACCCACGGTAAAAATCCTGTCCAAACAGTTCAATGACAAGGATGTGGATTTCTCGCACGTCGACTACGGTAAATGGGAGAGGAAGCCCATTAGATGTGCGTCCATGTCCAGAGTTGGGCTTAGAGGCCCAATTGAAATAAGGCCTGGAGAGTCATGGGCTTCTAGAAGTACAATAGGAGGCGGCCATTCAGATGCGGACTCCGAGATAGAGAACGAGCTCCATCCGTATAGACACATAAGTAAGCTCGGAACAGAAGTCCTAGATCCAGGAGAATCTGCTTCTATAGTAGGAGCCCAAAGAGCGGAATCCAATATAACTATGTCAATGGGGCAGTTAAACGAATTAGTTAGGACTACTGTACAAGAATGTATTAAAAGTAACTGTAAGGCTGTTCAGCCGAAATCATTGAAATAA |
| Protein Sequence | MESQLAFPPNAFNYIESHRDEYQLSHDLTEIILQFPSTASQLTARLSRSCMKIDHCVIEYRQQVPINATGSVVVEIHDKRMTDNESLQASWTFPIRCNIDLHYFSASFFSLKDPIPWKLYYRVCDTNVHQRTHFAKFKGKLKLSTAKHSVDIPFRAPTVKILSKQFNDKDVDFSHVDYGKWERKPIRCASMSRVGLRGPIEIRPGESWASRSTIGGGHSDADSEIENELHPYRHISKLGTEVLDPGESASIVGAQRAESNITMSMGQLNELVRTTVQECIKSNCKAVQPKSLK |
| NCBI Accession | YP_001294914.1 |
|---|---|
| Location | 196-951 |
| Gene Name | AV1 |
| Protein Name | coat protein |
| Coding Region | ATGCCTAAGCGTGATGCCCCATGGCGCTCGTTCGCGGCAACTTCCAAGGTTAGTCGCAATGCTAATTATTCTCCACGGACTGGTATGGGGCCAAAGGTCGACAAGGCCACTGCTTGGGTGAATAGGCCCATGTATAGAAAGCCCAGGATCTACCGGGTTATGAGAGGCCCAGATGTTCCAAAAGGATGTGAAGGCCCATGTAAGGTCCAGTCATACGAGCAGCGCCACGATATTTCACATGTTGGGAAGGTCATGTGTATCTCCGACGTGACACGTGGTAATGGTCTCACCCACCGTGTGGGGAAGCGTTTTTGTGTTAAGTCCGTGTATATTCTCGGCAAGGTTTGGATGGATGATAACATCAAGTTGAAGAACCACACGAACAGTGTTATGTTTTGGTTGGTCAGGGACCGTAGGCCTTATGGTATTCCTATGGATTTCGGACAAGTGTTTAACATGTTCGATAACGAACCCAGTACCGCGACGATCAAGAACGATCTTAGAGATCGGTTTCAGGTCATGCACAGGTTTTATGCTAAGGTTACAGGTGGACAGTATGCTAGTAACGAGCAAGCTATAGTGAGGCGTTTTTGGAAGGTCAACAATCATGTGGTCTACAATCATCAAGAGGCAGGAAAATATGACAACCATACTGAGAACGCCCTGTTATTGTATATGGCATGTACTCATGCTTCTAACCCCGTGTATGCAACATTGAAAATTCGGATCTATTTTTATGATTCGATTGCTAATTAA |
| Protein Sequence | MPKRDAPWRSFAATSKVSRNANYSPRTGMGPKVDKATAWVNRPMYRKPRIYRVMRGPDVPKGCEGPCKVQSYEQRHDISHVGKVMCISDVTRGNGLTHRVGKRFCVKSVYILGKVWMDDNIKLKNHTNSVMFWLVRDRRPYGIPMDFGQVFNMFDNEPSTATIKNDLRDRFQVMHRFYAKVTGGQYASNEQAIVRRFWKVNNHVVYNHQEAGKYDNHTENALLLYMACTHASNPVYATLKIRIYFYDSIAN |
| NCBI Accession | YP_001294915.1 |
|---|---|
| Location | 948-1346 |
| Gene Name | AC3 |
| Protein Name | replication enhancer |
| Coding Region | ATGGATTCACGCACCGGGGCGCTCATCACTGCACCTCAAGCAGAGAATGGCGTTTATATCTGGGAGATAGAAAATCCCCTCTATTTCAAGATGTACCGAGAAGAGGACATAGTGTACACGAGGACCAGGGTATACCATGTACAGATACGGTTCAACCACAACCTGAGGAGAGTGTTGCATCTCCACAAAGCATACCTGAACTTCCAAATCTGGACGACATTGATAACAGCTTCTGGGTCGAATTATTTAGCTAGGTTTAGACATTTAGTTAATATGTATTTAGATCAGTTAGGTGTGATTTCGATAAACAATGTAATCAGAGCTGTTCGTTTTGCAACAGACAGATCATATGTAAATTATGTACTTGAAGATCATTCAATAAAAATAAAACTTTATTAA |
| Protein Sequence | MDSRTGALITAPQAENGVYIWEIENPLYFKMYREEDIVYTRTRVYHVQIRFNHNLRRVLHLHKAYLNFQIWTTLITASGSNYLARFRHLVNMYLDQLGVISINNVIRAVRFATDRSYVNYVLEDHSIKIKLY |
| NCBI Accession | YP_001294916.1 |
|---|---|
| Location | 1093-1482 |
| Gene Name | AC2 |
| Protein Name | transcriptional activator |
| Coding Region | ATGCAATCTTCGTCACCCTCCAATCCGCCCTCTATCAAGACAGCACACAGGCAGGCCAAGAAGAGGAGCATTAGAAGAAGGCGGATTGATCTAGAGTGCGGGTGCACCATCTACCTCCACATAGACTGCACGGGACATGGATTCACGCACCGGGGCGCTCATCACTGCACCTCAAGCAGAGAATGGCGTTTATATCTGGGAGATAGAAAATCCCCTCTATTTCAAGATGTACCGAGAAGAGGACATAGTGTACACGAGGACCAGGGTATACCATGTACAGATACGGTTCAACCACAACCTGAGGAGAGTGTTGCATCTCCACAAAGCATACCTGAACTTCCAAATCTGGACGACATTGATAACAGCTTCTGGGTCGAATTATTTAGCTAG |
| Protein Sequence | MQSSSPSNPPSIKTAHRQAKKRSIRRRRIDLECGCTIYLHIDCTGHGFTHRGAHHCTSSREWRLYLGDRKSPLFQDVPRRGHSVHEDQGIPCTDTVQPQPEESVASPQSIPELPNLDDIDNSFWVELFS |
| NCBI Accession | YP_001294917.1 |
|---|---|
| Location | 1409-2479 |
| Gene Name | AC1 |
| Protein Name | replication protein |
| Coding Region | ATGCCATCGGTTAAACGCTTCAAAGTCTCAGCTAAAAACTATTTTCTCACATATCCCCAGTGTTCTCTTACAAAAGAAGAAGCACTTTCCCAATTACAAAACCTAACAATTCCGGTTAACAAGAAATTCATCAAGATCTGCAGAGAGCTCCACGAGAATGGGGAACCTCATCTCCACGTCCTTATCCAGTTCGAAGGCAAATACCAGTGCACGAATAACAGATTCTTCGATCTGGTGTCCCCTACCAGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAATCAAGTTCCGACGTCAAGTCCTACATCGACAAGGATGGAGATACAATCGAATGGGGACAATTCCAGATCGACGGTAGATCTGCCAGAGGAGGTAAACAATCTACAAACGACACATATGCCAAGGCGTTGAATGCAGCATGTGCAGACGAAGCAATGCGAATCATACGGGAAGAACAACCACAACATTTCTTCCTTCAACATCATAACCTAGTCGCTAACGCGGCTAAAATATTCAAAAAGGCTCCGGAACGCTGGGTTCCTCCGTTTCAACTCTCTTCTTTCACTAACGTTCCGGACGAGATGCAAGAATGGGCAGATGATTATTTTGGAAAGGATTCAGCTGCGCGGCCAGAGAGACCTATTAGTATTATCATAGAAGGTGATTCTAGGACAGGGAAGACGATGTGGGCTCGGGCATTAGGCCCACATAATTATCTCAGTGGACATCTCGACTTCAATTCCAGAGTGTTCTCTAATGAAGTGGAGTATAACGTCATTGACGACGTCACACCGCAATATCTAAAGCTAAAGCACTGGAAAGAACTTCTGGGGGCCCAGAAAGACTGGCAGTCAAATTGCAAGTATGGCAAGCCAGTTCAAATTAAAGGTGGGATCCCATCAATCGTGCTTTGCAATCCTGGTGAGGGTGCCAGCTATAAAGATTACCTAAACAAAGAAGAAAACACATCACTCAAAAATTGGACACTCAAGAATGCAATCTTCGTCACCCTCCAATCCGCCCTCTATCAAGACAGCACACAGGCAGGCCAAGAAGAGGAGCATTAG |
| Protein Sequence | MPSVKRFKVSAKNYFLTYPQCSLTKEEALSQLQNLTIPVNKKFIKICRELHENGEPHLHVLIQFEGKYQCTNNRFFDLVSPTRSAHFHPNIQGAKSSSDVKSYIDKDGDTIEWGQFQIDGRSARGGKQSTNDTYAKALNAACADEAMRIIREEQPQHFFLQHHNLVANAAKIFKKAPERWVPPFQLSSFTNVPDEMQEWADDYFGKDSAARPERPISIIIEGDSRTGKTMWARALGPHNYLSGHLDFNSRVFSNEVEYNVIDDVTPQYLKLKHWKELLGAQKDWQSNCKYGKPVQIKGGIPSIVLCNPGEGASYKDYLNKEENTSLKNWTLKNAIFVTLQSALYQDSTQAGQEEEH |
| NCBI Accession | YP_001294918.1 |
|---|---|
| Location | 2065-2322 |
| Gene Name | AC4 |
| Protein Name | AC4 protein |
| Coding Region | ATGGGGAACCTCATCTCCACGTCCTTATCCAGTTCGAAGGCAAATACCAGTGCACGAATAACAGATTCTTCGATCTGGTGTCCCCTACCAGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAATCAAGTTCCGACGTCAAGTCCTACATCGACAAGGATGGAGATACAATCGAATGGGGACAATTCCAGATCGACGGTAGATCTGCCAGAGGAGGTAAACAATCTACAAACGACACATATGCCAAGGCGTTGA |
| Protein Sequence | MGNLISTSLSSSKANTSARITDSSIWCPLPGQHISIRTFRELNQVPTSSPTSTRMEIQSNGDNSRSTVDLPEEVNNLQTTHMPRR |
References More References in PubMed
| 1 |
First Report of Sinaloa Tomato Leaf Curl Geminivirus in Costa Rica. Idris AM, et al. Plant Dis. 1999 Mar;83(3):303. doi: 10.1094/PDIS.1999.83.3.303C. PMID: 30845524 |
|---|---|
| 2 |
First Report of Tomato yellow leaf curl virus in Tomato in Costa Rica. Barboza N, et al. Plant Dis. 2014 May;98(5):699. doi: 10.1094/PDIS-08-13-0881-PDN. PMID: 30708517 |
| 3 |
Idris AM, et al. Phytopathology. 1998 Jul;88(7):648-57. doi: 10.1094/PHYTO.1998.88.7.648. PMID: 18944936 |
| 4 |
Gámez-Jiménez C, et al. Plant Dis. 2009 May;93(5):545. doi: 10.1094/PDIS-93-5-0545A. PMID: 30764171 |
| 5 |
Rojas A, et al. Arch Virol. 2005 Jul;150(7):1281-99. doi: 10.1007/s00705-005-0509-x. Epub 2005 Mar 24. PMID: 15789265 |
| 6 |
Introduction of the Exotic Monopartite Tomato yellow leaf curl virus into West Coast Mexico. Brown JK, et al. Plant Dis. 2006 Oct;90(10):1360. doi: 10.1094/PD-90-1360A. PMID: 30780952 |
| 7 |
Rojas MR, et al. Plant Dis. 2007 Aug;91(8):1056. doi: 10.1094/PDIS-91-8-1056A. PMID: 30780456 |
| 8 |
A New Begomovirus Causes Tomato Leaf Curl Disease in Baja California Sur, Mexico. Holguín-Peña RJ, et al. Plant Dis. 2005 Mar;89(3):341. doi: 10.1094/PD-89-0341A. PMID: 30795368 |
| 9 |
Idris AM, et al. Plant Dis. 2007 Jul;91(7):910. doi: 10.1094/PDIS-91-7-0910C. PMID: 30780421 |
| 10 |
Mendoza-Figueroa JS, et al. Pestic Biochem Physiol. 2018 Feb;145:56-65. doi: 10.1016/j.pestbp.2018.01.005. Epub 2018 Feb 4. PMID: 29482732 |