Tomato leaf curl New Delhi virus 5
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_004786775.1 |
| Isolate | Bangladesh |
| Release date | 2021/6/1 |
| Submitter | Monger,W.A., Mumford,R., Kelly,P. |
| Host | |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
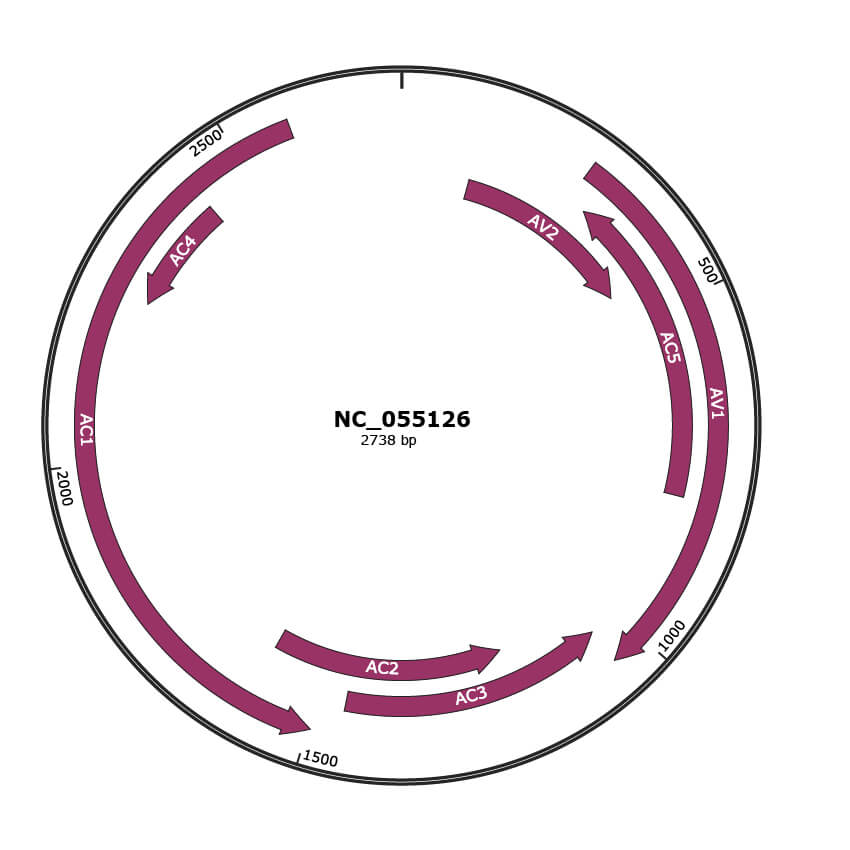
JBrowse
Genome
NC_055126
Gene Information
| NCBI Accession | YP_010084316.1 |
|---|---|
| Location | 118-447 |
| Gene Name | AV2 |
| Protein Name | pre-coat protein |
| Coding Region | ATGTGGGATCCATTATTGCACGAATTTCCTGAAAGCGTGCATGGTCTAAGGTGCATGCTAGCTGTAAAATATCTTCAAGAGATAGAGAAGAACTATTCCCCAGACACAGTGGGCTACGATCTTGTCCGAGATCTAATTTTAGTAGTTCGAGCAAAGAACTATGTCGAAGCGACCAGCAGATATCATCATTTCAACTCCCGCATCGAAGGTACGCCGACGTCTCAACTTCGACAGCCCTTATGGAGCTCGTGCAGTTGTCCCCATTGCCCGCGTCACAAAAGCAAAGGCGTGGACAAACAGGCCGATGAACAGAAAGCCCAGAATGTATAG |
| Protein Sequence | MWDPLLHEFPESVHGLRCMLAVKYLQEIEKNYSPDTVGYDLVRDLILVVRAKNYVEATSRYHHFNSRIEGTPTSQLRQPLWSSCSCPHCPRHKSKGVDKQADEQKAQNV |
| NCBI Accession | YP_010084317.1 |
|---|---|
| Location | 278-1048 |
| Gene Name | AV1 |
| Protein Name | coat protein |
| Coding Region | ATGTCGAAGCGACCAGCAGATATCATCATTTCAACTCCCGCATCGAAGGTACGCCGACGTCTCAACTTCGACAGCCCTTATGGAGCTCGTGCAGTTGTCCCCATTGCCCGCGTCACAAAAGCAAAGGCGTGGACAAACAGGCCGATGAACAGAAAGCCCAGAATGTATAGAATGTACAGAAGCCCCGACGTGCCAAGGGGATGTGAAGGTCCTTGTAAGGTGCAGTCTTTTGAATCTAGGCACGATGTGTCTCATATTGGCAAAGTCATGTGTGTCAGTGACGTTACCCGAGGAACCGGACTGACTCATCGCGTGGGGAAGCGTTTCTGTGTGAAATCTGTGTATGTATTGGGGAAGATATGGATGGATGAAAACATAAAGACTAAAAACCACACGAACAGTGTGATGTTTTTCTTGGTTCGTGACCGTCGTCCTACGGGATCGCCACAGGATTTTGGTGAAGTGTTCAACATGTTTGACAATGAACCGAGCACCGCAACGGTGAAGAACATGCATCGTGATCGTTATCAAGTGTTAAGGAAATGGCATGCGACTGTGACTGGAGGGACTTATGCATCGAAGGAGCAAGCATTAGTTAGGAAGTTTGTTAGGGTTAATAATTATGTCGTTTATAATCAACAAGAGGCCGGAAAGTATGAAAATCATACTGAGAACGCATTAATGTTGTATATGGCCTGTACTCATGCATCAAATCCTGTGTATGCTACTTTGAAAATCCGGATCTATTTTTATGATTCGGTAACAAATTAA |
| Protein Sequence | MSKRPADIIISTPASKVRRRLNFDSPYGARAVVPIARVTKAKAWTNRPMNRKPRMYRMYRSPDVPRGCEGPCKVQSFESRHDVSHIGKVMCVSDVTRGTGLTHRVGKRFCVKSVYVLGKIWMDENIKTKNHTNSVMFFLVRDRRPTGSPQDFGEVFNMFDNEPSTATVKNMHRDRYQVLRKWHATVTGGTYASKEQALVRKFVRVNNYVVYNQQEAGKYENHTENALMLYMACTHASNPVYATLKIRIYFYDSVTN |
| NCBI Accession | YP_010084318.1 |
|---|---|
| Location | 308-793 |
| Gene Name | AC5 |
| Protein Name | AC5 |
| Coding Region | ATGCATGTTCTTCACCGTTGCGGTGCTCGGTTCATTGTCAAACATGTTGAACACTTCACCAAAATCCTGTGGCGATCCCGTAGGACGACGGTCACGAACCAAGAAAAACATCACACTGTTCGTGTGGTTTTTAGTCTTTATGTTTTCATCCATCCATATCTTCCCCAATACATACACAGATTTCACACAGAAACGCTTCCCCACGCGATGAGTCAGTCCGGTTCCTCGGGTAACGTCACTGACACACATGACTTTGCCAATATGAGACACATCGTGCCTAGATTCAAAAGACTGCACCTTACAAGGACCTTCACATCCCCTTGGCACGTCGGGGCTTCTGTACATTCTATACATTCTGGGCTTTCTGTTCATCGGCCTGTTTGTCCACGCCTTTGCTTTTGTGACGCGGGCAATGGGGACAACTGCACGAGCTCCATAAGGGCTGTCGAAGTTGAGACGTCGGCGTACCTTCGATGCGGGAGTTGA |
| Protein Sequence | MHVLHRCGARFIVKHVEHFTKILWRSRRTTVTNQEKHHTVRVVFSLYVFIHPYLPQYIHRFHTETLPHAMSQSGSSGNVTDTHDFANMRHIVPRFKRLHLTRTFTSPWHVGASVHSIHSGLSVHRPVCPRLCFCDAGNGDNCTSSIRAVEVETSAYLRCGS |
| NCBI Accession | YP_010084319.1 |
|---|---|
| Location | 1045-1455 |
| Gene Name | AC3 |
| Protein Name | replication enhancer protein |
| Coding Region | ATGATCACGGATTCACGCACAGGGGAGTACATCACTGCGGATCGAGTAGAGAGTGGCGTGTTTATTTGGGACGTTCCAAATCCCCTGTATTTCAAAATATTGAAACACGACAGCAGACCATGCCTCACGAATCACGACATCATCCACCTCCAAATACAGTTCAACCACAACCTGCGGAAAGCTCTGGGGCTTCACAAGTGTTTTCTAATTTTCCAAATCTGGACGGGCTTACGCCCTCCGACTGGGATTTTCTTGAGGGTATTTAAGACACAGGTCCTTAAATACCTCCATGAGCTTGGTGTAATAGGAATTTCGAATGTAATAAGAGCAGCCTATCATGTATTGGATAATGTTTTAGAAAAGACTATTGATGTATTGCCATCATGTGATGTAAGGATTAATATATATTAA |
| Protein Sequence | MITDSRTGEYITADRVESGVFIWDVPNPLYFKILKHDSRPCLTNHDIIHLQIQFNHNLRKALGLHKCFLIFQIWTGLRPPTGIFLRVFKTQVLKYLHELGVIGISNVIRAAYHVLDNVLEKTIDVLPSCDVRINIY |
| NCBI Accession | YP_010084320.1 |
|---|---|
| Location | 1190-1594 |
| Gene Name | AC2 |
| Protein Name | transcriptional activator protein |
| Coding Region | ATGCAGTCTTCGTCACACTCCAAGAACCACTCTATTCCGGTCGCGAAAACATCAGTCCCCCAGAAGAAGAAGAAGAACATACGCAGGAGGCGAGTTGATCTGCAGTGTGGTTGTTCGTACTACATATCAATCAACTGTCATGATCACGGATTCACGCACAGGGGAGTACATCACTGCGGATCGAGTAGAGAGTGGCGTGTTTATTTGGGACGTTCCAAATCCCCTGTATTTCAAAATATTGAAACACGACAGCAGACCATGCCTCACGAATCACGACATCATCCACCTCCAAATACAGTTCAACCACAACCTGCGGAAAGCTCTGGGGCTTCACAAGTGTTTTCTAATTTTCCAAATCTGGACGGGCTTACGCCCTCCGACTGGGATTTTCTTGAGGGTATTTAA |
| Protein Sequence | MQSSSHSKNHSIPVAKTSVPQKKKKNIRRRRVDLQCGCSYYISINCHDHGFTHRGVHHCGSSREWRVYLGRSKSPVFQNIETRQQTMPHESRHHPPPNTVQPQPAESSGASQVFSNFPNLDGLTPSDWDFLEGI |
| NCBI Accession | YP_010084321.1 |
|---|---|
| Location | 1497-2582 |
| Gene Name | AC1 |
| Protein Name | replication initiator protein |
| Coding Region | ATGGCTCCGCCAAGTCGTTTTAGAATTAATGCCAAAAACTATTTCCTCACTTACCCAAAATGCTCTCTTACAAAGGAAGAGGCACTTTCCCAATTAATAAACCTAGAAACCCCTACTTGCAAGAAATTCATCAAAATCTGTAGAGAGAGTCACGAGGATGGGTCTCCGCATATCCATGTTCTCATTCAATTCGAAGGGAAATACCAGTGCAAGAATAACAGATTCTTCGACTTGGTATCCCCAACTCGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAATCGGCGTCCGACGTCAAGGCATATATCGACAAAGACGGAGACGTGCTAGAATGGGGTGTTTTCCAGATCGACGGACGATCTGCTCGTGGGGGTCAACAGACGGCAAACGATGCATACGCTCAGGCAATTAACACCGGAAACAAAGAAGATGCATTGAAAGTACTAAAAGAACTAGCACCAAAAGATTATGTTCTGCAGTTCCATAATTTAATGACGAATTTAGATCGCATTTTTCAACCACGTTCTGAGGTCTATGTATCTCCATTCTCAACTACTTCCTTCGACACAGTTCCTCCAGAACTCGTCGATTGGGTTCAGTCCAATGTATTGTCTGCCGCTGCGCGGCCTTTTAGACCAATAAGCGTAGTAATAGAGGGGGATAGTAGAACGGGAAAGACAATGTGGGCACGTTCACTAGGACCCCACAATTACTTGTGTGGTCATCTGGATCTCAGCCCGAAAGTGTACAATAATGAAGCGTGGTACAACGTCATTGATGACGTCGATCCCCACTATCTAAAGCACTTTAAAGAATTCATGGGGGCCCAGCGTGACTGGCAAAGCAATACGAAGTACGGAAAGCCGGTCATGATTAAAGGCGGTATTCCCACCATCTTCCTCTGCAATAAAGGACCAAACAGCAGCTATAAGGAATATCTCGACGAAGAAAAAAATGCATCACTGAAACAGTGGGCTATCAAGAATGCAGTCTTCGTCACACTCCAAGAACCACTCTATTCCGGTCGCGAAAACATCAGTCCCCCAGAAGAAGAAGAAGAACATACGCAGGAGGCGAGTTGA |
| Protein Sequence | MAPPSRFRINAKNYFLTYPKCSLTKEEALSQLINLETPTCKKFIKICRESHEDGSPHIHVLIQFEGKYQCKNNRFFDLVSPTRSAHFHPNIQGAKSASDVKAYIDKDGDVLEWGVFQIDGRSARGGQQTANDAYAQAINTGNKEDALKVLKELAPKDYVLQFHNLMTNLDRIFQPRSEVYVSPFSTTSFDTVPPELVDWVQSNVLSAAARPFRPISVVIEGDSRTGKTMWARSLGPHNYLCGHLDLSPKVYNNEAWYNVIDDVDPHYLKHFKEFMGAQRDWQSNTKYGKPVMIKGGIPTIFLCNKGPNSSYKEYLDEEKNASLKQWAIKNAVFVTLQEPLYSGRENISPPEEEEEHTQEAS |
| NCBI Accession | YP_010084322.1 |
|---|---|
| Location | 2249-2425 |
| Gene Name | AC4 |
| Protein Name | AC4 |
| Coding Region | ATGGGTCTCCGCATATCCATGTTCTCATTCAATTCGAAGGGAAATACCAGTGCAAGAATAACAGATTCTTCGACTTGGTATCCCCAACTCGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAATCGGCGTCCGACGTCAAGGCATATATCGACAAAGACGGAGACGTGCTAG |
| Protein Sequence | MGLRISMFSFNSKGNTSARITDSSTWYPQLGQHISIRTFRELNRRPTSRHISTKTETC |
References More References in PubMed
| 1 |
Naganur P, et al. Plants (Basel). 2023 Jan 20;12(3):490. doi: 10.3390/plants12030490. PMID: 36771575 |
|---|---|
| 2 |
Seed Transmission of Tomato Leaf Curl New Delhi Virus from Zucchini Squash in Italy. Kil EJ, et al. Plants (Basel). 2020 Apr 29;9(5):563. doi: 10.3390/plants9050563. PMID: 32365497 |
| 3 |
Jeevalatha A, et al. 3 Biotech. 2021 Apr;11(4):203. doi: 10.1007/s13205-021-02752-5. Epub 2021 Mar 30. PMID: 33927993 |
| 4 |
Venkataravanappa V, et al. 3 Biotech. 2020 Jun;10(6):282. doi: 10.1007/s13205-020-02245-x. Epub 2020 Jun 2. PMID: 32550101 |
| 5 |
Prasad A, et al. Theor Appl Genet. 2021 May;134(5):1463-1474. doi: 10.1007/s00122-021-03783-5. Epub 2021 Feb 7. PMID: 33554270 |
| 6 |
Qiao N, et al. Plant Dis. 2025 Sep 18. doi: 10.1094/PDIS-07-25-1386-RE. Online ahead of print. PMID: 40968395 |
| 7 |
Naga KC, et al. 3 Biotech. 2021 Sep;11(9):421. doi: 10.1007/s13205-021-02966-7. Epub 2021 Aug 31. PMID: 34603921 |
| 8 |
Nagendran K, et al. Virusdisease. 2017 Dec;28(4):425-429. doi: 10.1007/s13337-017-0403-7. Epub 2017 Oct 9. PMID: 29291235 |
| 9 |
First Report of Tomato leaf curl New Delhi virus Infecting Cucumber in Central Java, Indonesia. Mizutani T, et al. Plant Dis. 2011 Nov;95(11):1485. doi: 10.1094/PDIS-03-11-0196. PMID: 30731770 |
| 10 |
Vo TTB, et al. Plants (Basel). 2022 Mar 6;11(5):704. doi: 10.3390/plants11050704. PMID: 35270174 |