Tomato leaf curl New Delhi virus 4
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_002824185.1 |
| Isolate | India: Gujarat, Junagad |
| Release date | 2018/8/25 |
| Submitter | Swarnalatha,P., Manasa,M., Venkataravanappa,V., Jalali,S., KrishnaReddy,M. |
| Host | |
| Download | Genome |GFF3 |PEP |CDS |
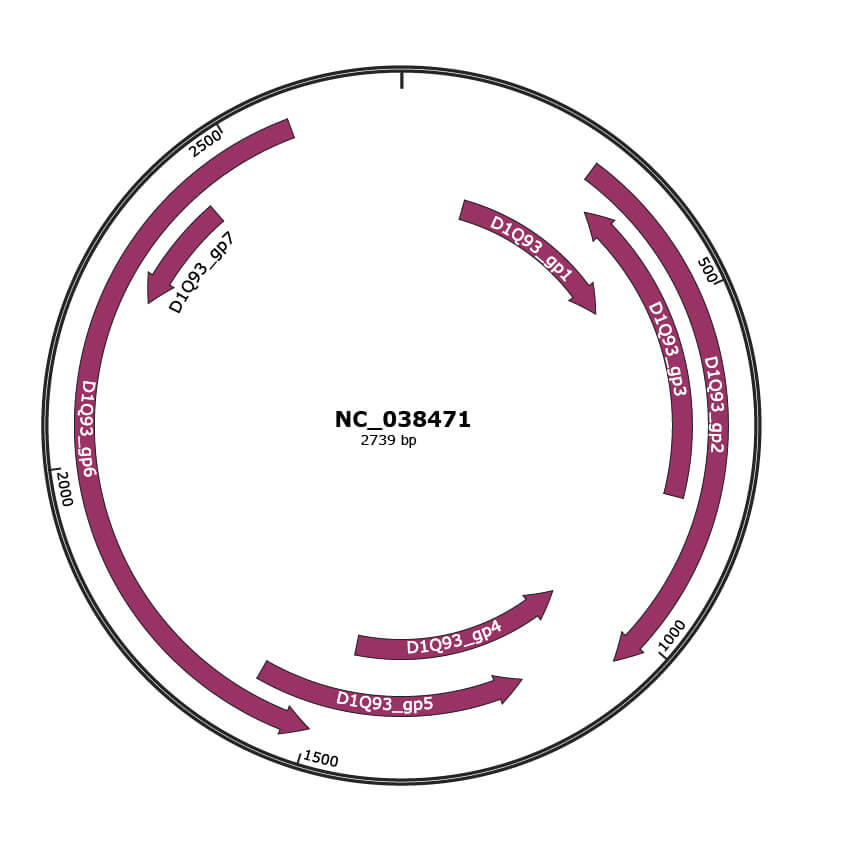
Genomic Organization
JBrowse
Genome
NC_038471
Gene Information
| NCBI Accession | YP_009506532.1 |
|---|---|
| Location | 120-458 |
| Protein Name | AV2 |
| Coding Region | ATGCGGGATCCATTATTGCTCGCCTTTCCAGAAAGAGTCCATGGTGTAAGGAGCATGTTAGTTGAAAAACCTCTCCAAGAGATGGAAGACCATTATTCACCAGACACAGTAGGCTACGATTTTATCCGAGATCTCATTCTTGTTCTTCGAGCAAAAAACTATGAAGAACCGACCAGCAGATATCATCATATCAAAGCCCCCATCGAAGGTATACAGATATGTCAAGTTCGACAGCCCCAAAGGAGCGAGTGCAGTGGTCCCCATTGCCCGCGTCACAGAAGCAAAGGCCTGGACCAACAGGCCTATGAACAGAAAACCCAGAATATACAGAACGCATAG |
| Protein Sequence | MRDPLLLAFPERVHGVRSMLVEKPLQEMEDHYSPDTVGYDFIRDLILVLRAKNYEEPTSRYHHIKAPIEGIQICQVRQPQRSECSGPHCPRHRSKGLDQQAYEQKTQNIQNA |
| NCBI Accession | YP_009506533.1 |
|---|---|
| Location | 280-1050 |
| Protein Name | AV1 |
| Coding Region | ATGAAGAACCGACCAGCAGATATCATCATATCAAAGCCCCCATCGAAGGTATACAGATATGTCAAGTTCGACAGCCCCAAAGGAGCGAGTGCAGTGGTCCCCATTGCCCGCGTCACAGAAGCAAAGGCCTGGACCAACAGGCCTATGAACAGAAAACCCAGAATATACAGAACGCATAGAAGTCCAGACGTGCCAAGGGGCCGTGAAGGCCAGAGGAAATTGCACTCGATTGAATCCAGGCTTGATGTATATCGGACTGGCAAAATCAAGGGTGTTATTCATGTTACCCGAGAAATCGGACTCACACATAGCGTAGGGAAGCGATTTGGGGTGAATTCGGTTTATGTGCTGGAAAAGAGAGGGATCGATGAAACCATCAAGCCAAAAAGCCATACTTACCGGGTAATGTTTTTTTTGGTTCGTGACCGTCGTCCTACAGGAACTCCCCAGGATTTCGGGGACGTGTTTAATATGTTTGACAATGAACCGAGCACAGCAACGGTGAAGAACATGCATCGTGATCGTTATCAAGTCTTACGGAAGTGGCATGCAACTGTGACCGGAGGAACATATGCATCTAAGGAGCAAGCATTAGTTAGCAAGTTTGTTAGGGTTAATAATTATGTAGTTTATAATCAACAAGAGGCCGGCAAGTATGAGAATCATACTGAAAACGCATTAATGTTGTATATGGCCTGCACTCATGCATCAAATCCTGTATATGCTACTTTGAAAATCCGGATCTATTTCTATGATTCGGTCACAAATTAA |
| Protein Sequence | MKNRPADIIISKPPSKVYRYVKFDSPKGASAVVPIARVTEAKAWTNRPMNRKPRIYRTHRSPDVPRGREGQRKLHSIESRLDVYRTGKIKGVIHVTREIGLTHSVGKRFGVNSVYVLEKRGIDETIKPKSHTYRVMFFLVRDRRPTGTPQDFGDVFNMFDNEPSTATVKNMHRDRYQVLRKWHATVTGGTYASKEQALVSKFVRVNNYVVYNQQEAGKYENHTENALMLYMACTHASNPVYATLKIRIYFYDSVTN |
| NCBI Accession | YP_009506534.1 |
|---|---|
| Location | 310-795 |
| Protein Name | AC5 |
| Coding Region | ATGCATGTTCTTCACCGTTGCTGTGCTCGGTTCATTGTCAAACATATTAAACACGTCCCCGAAATCCTGGGGAGTTCCTGTAGGACGACGGTCACGAACCAAAAAAAACATTACCCGGTAAGTATGGCTTTTTGGCTTGATGGTTTCATCGATCCCTCTCTTTTCCAGCACATAAACCGAATTCACCCCAAATCGCTTCCCTACGCTATGTGTGAGTCCGATTTCTCGGGTAACATGAATAACACCCTTGATTTTGCCAGTCCGATATACATCAAGCCTGGATTCAATCGAGTGCAATTTCCTCTGGCCTTCACGGCCCCTTGGCACGTCTGGACTTCTATGCGTTCTGTATATTCTGGGTTTTCTGTTCATAGGCCTGTTGGTCCAGGCCTTTGCTTCTGTGACGCGGGCAATGGGGACCACTGCACTCGCTCCTTTGGGGCTGTCGAACTTGACATATCTGTATACCTTCGATGGGGGCTTTGA |
| Protein Sequence | MHVLHRCCARFIVKHIKHVPEILGSSCRTTVTNQKKHYPVSMAFWLDGFIDPSLFQHINRIHPKSLPYAMCESDFSGNMNNTLDFASPIYIKPGFNRVQFPLAFTAPWHVWTSMRSVYSGFSVHRPVGPGLCFCDAGNGDHCTRSFGAVELDISVYLRWGL |
| NCBI Accession | YP_009506535.1 |
|---|---|
| Location | 1047-1457 |
| Protein Name | AC3 |
| Coding Region | ATGACCACGGATTCACGCACAGGGGAGTACATCACTGCGGATCGTACAGAGAGTGGCGTATATACCTGGGAAATCAGAAATCCCCTGTATTTCAAAATTTTGAAACACGACAGCAGGCCCTTCCTAACACAGCACGACATAATCAAGATTCAAATACAATTCAACCACAACCTGCGGAAAGCCCTGGGACTTCACAAGTGTTTTCTAACTTTCAAAATCTGGACGGGCTTACGCCCTCAGATTGGGATTTTCTTGAGAGTCTTCAAAACCCAGGTCCTTAAGTACCTGGATATGTTAGGTGTAATAGGAATTAATAATGTAATAAGAGCAGTTTATCATGTTTTGGATAATGTAATGGAGCGAACAATTGATGTATGGACTACATATGATATAAAGCTTGATATTTATTAA |
| Protein Sequence | MTTDSRTGEYITADRTESGVYTWEIRNPLYFKILKHDSRPFLTQHDIIKIQIQFNHNLRKALGLHKCFLTFKIWTGLRPQIGIFLRVFKTQVLKYLDMLGVIGINNVIRAVYHVLDNVMERTIDVWTTYDIKLDIY |
| NCBI Accession | YP_009506536.1 |
|---|---|
| Location | 1177-1596 |
| Protein Name | AC2 |
| Coding Region | ATGCGACCTTCGTCACCCTCGAAGGCCCACTCTACTCAGGTACCAATCAAAGTACAGCACAAGCTAGCCAAGAAGGGGACCAGACGTCGACGAGTTGATCTACCGTGTGGCTGTTCTTACTTCATAGCATTAACCTGCCATGACCACGGATTCACGCACAGGGGAGTACATCACTGCGGATCGTACAGAGAGTGGCGTATATACCTGGGAAATCAGAAATCCCCTGTATTTCAAAATTTTGAAACACGACAGCAGGCCCTTCCTAACACAGCACGACATAATCAAGATTCAAATACAATTCAACCACAACCTGCGGAAAGCCCTGGGACTTCACAAGTGTTTTCTAACTTTCAAAATCTGGACGGGCTTACGCCCTCAGATTGGGATTTTCTTGAGAGTCTTCAAAACCCAGGTCCTTAA |
| Protein Sequence | MRPSSPSKAHSTQVPIKVQHKLAKKGTRRRRVDLPCGCSYFIALTCHDHGFTHRGVHHCGSYREWRIYLGNQKSPVFQNFETRQQALPNTARHNQDSNTIQPQPAESPGTSQVFSNFQNLDGLTPSDWDFLESLQNPGP |
| NCBI Accession | YP_009506537.1 |
|---|---|
| Location | 1499-2584 |
| Protein Name | AC1 |
| Coding Region | ATGGTTTCGCCAGGTCGTGTTAGGGAAAATGCCAAAAAGTATTTCCTCACAGATCCAAAGTGCTCTCTAACTAAAGGAGAGGCACTTTGTCAATTGCAAACCAGGGAAACCCAGGGGCAGAAGAAATTCCTCTCGATCTGTAGAGAGCTTCATGTAGATGGGTTTCGGCATATCCAGGTTGTCATCCAATTCGAAGGGAAATTCCACTGCAAGAATAACAGATGCTTGGAATTGGTTTCCCCAAGTCGGTCAGCACATTTCCATCCGGACATTCAGGGAGCTAAGAGCTCAAGCGATGTCAAATCCTACATGGAAAAAGACGGAGACGTTCTAGAATGGGGTGTTTTCCAAGTCGATGGACGATCAGCTAGAGGAGGTTGCCAATCTGCCAACGACCCATATGCCGAGGCAATTAATTCAGGGTCCAAATCTCAGGCCCTCAATATATTAAGGGAGAAATCTCCTACATATTTCGTTTTACAATTTCACAATTTAAATACCAATTTAGATCGCATTTTTCAACCTCCTTCTGAGGTTTATGTTTCTCCATTTTCAATTTCATCCTTCGACAGAGTTCCGGCAGACCTCGTCGATTGGGTTACGTCAAATGTTGTGTGTGCCGCTGCGCGGCCTTTTAGGCCCATAAGCATAGTCATAGAGGGGGATAGTAGAACTGGCAAAACAATGTGGGCTCGTTGCTTAGGTCCACATAATTATTTGTGTGGTCATTTTGACCTTAGGCCAAAGGTGTACAGTAATGAAGCCTGGTACAACGTCATTGATGACGTCGACCCCCACTATCTAAAGCACTTTAAAGAAATCATGGGGGCCCAAAGGGACTGGCAATCAAATACAAAGTACGGGAAGCCAGTTCAAATTAAAGGCGGGAATCCCACTACTTTCCTCTGCAATCCAGGACCCAACTCCAGCTATAAAGAATACTTGGATGAGGAAAAGAATTCTGCACTAAAAACCTGGGCTTTAAAAAATGCGACCTTCGTCACCCTCGAAGGCCCACTCTACTCAGGTACCAATCAAAGTACAGCACAAGCTAGCCAAGAAGGGGACCAGACGTCGACGAGTTGA |
| Protein Sequence | MVSPGRVRENAKKYFLTDPKCSLTKGEALCQLQTRETQGQKKFLSICRELHVDGFRHIQVVIQFEGKFHCKNNRCLELVSPSRSAHFHPDIQGAKSSSDVKSYMEKDGDVLEWGVFQVDGRSARGGCQSANDPYAEAINSGSKSQALNILREKSPTYFVLQFHNLNTNLDRIFQPPSEVYVSPFSISSFDRVPADLVDWVTSNVVCAAARPFRPISIVIEGDSRTGKTMWARCLGPHNYLCGHFDLRPKVYSNEAWYNVIDDVDPHYLKHFKEIMGAQRDWQSNTKYGKPVQIKGGNPTTFLCNPGPNSSYKEYLDEEKNSALKTWALKNATFVTLEGPLYSGTNQSTAQASQEGDQTSTS |
| NCBI Accession | YP_009506538.1 |
|---|---|
| Location | 2251-2427 |
| Protein Name | AC4 |
| Coding Region | ATGGGTTTCGGCATATCCAGGTTGTCATCCAATTCGAAGGGAAATTCCACTGCAAGAATAACAGATGCTTGGAATTGGTTTCCCCAAGTCGGTCAGCACATTTCCATCCGGACATTCAGGGAGCTAAGAGCTCAAGCGATGTCAAATCCTACATGGAAAAAGACGGAGACGTTCTAG |
| Protein Sequence | MGFGISRLSSNSKGNSTARITDAWNWFPQVGQHISIRTFRELRAQAMSNPTWKKTETF |
References More References in PubMed
| 1 |
Tomato leaf curl New Delhi virus: an emerging plant begomovirus threatening cucurbit production. Cai L, et al. aBIOTECH. 2023 Oct 25;4(3):257-266. doi: 10.1007/s42994-023-00118-4. eCollection 2023 Sep. PMID: 37970471 |
|---|---|
| 2 |
Farina A, et al. Insects. 2023 Apr 15;14(4):384. doi: 10.3390/insects14040384. PMID: 37103199 |
| 3 |
Whitefly Control Strategies against Tomato Leaf Curl New Delhi Virus in Greenhouse Zucchini. Rodríguez E, et al. Int J Environ Res Public Health. 2019 Jul 26;16(15):2673. doi: 10.3390/ijerph16152673. PMID: 31357394 |
| 4 |
Taglienti A, et al. Front Microbiol. 2022 Apr 25;13:840893. doi: 10.3389/fmicb.2022.840893. eCollection 2022. PMID: 35547120 |
| 5 |
Jeevalatha A, et al. 3 Biotech. 2021 Apr;11(4):203. doi: 10.1007/s13205-021-02752-5. Epub 2021 Mar 30. PMID: 33927993 |
| 6 |
Resistance Against Melon Chlorotic Mosaic Virus and Tomato Leaf Curl New Delhi Virus in Melon. Romay G, et al. Plant Dis. 2019 Nov;103(11):2913-2919. doi: 10.1094/PDIS-02-19-0298-RE. Epub 2019 Aug 20. PMID: 31436474 |
| 7 |
Prasad A, et al. Theor Appl Genet. 2021 May;134(5):1463-1474. doi: 10.1007/s00122-021-03783-5. Epub 2021 Feb 7. PMID: 33554270 |
| 8 |
Chang HH, et al. Mol Plant Pathol. 2022 Apr;23(4):561-575. doi: 10.1111/mpp.13181. Epub 2022 Jan 4. PMID: 34984809 |
| 9 |
Venkataravanappa V, et al. Iran J Biotechnol. 2019 Jan 11;17(1):e2134. doi: 10.21859/ijb.2134. eCollection 2019 Jan. PMID: 31457044 |
| 10 |
A distinct seed-transmissible strain of tomato leaf curl New Delhi virus infecting Chayote in India. Sangeetha B, et al. Virus Res. 2018 Oct 15;258:81-91. doi: 10.1016/j.virusres.2018.10.009. Epub 2018 Oct 15. PMID: 30336187 |