Tomato leaf curl Karnataka virus 3
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_004787135.1 |
| Isolate | India: Rauke, Punjab |
| Release date | 2021/6/1 |
| Submitter | Swarnalatha,P., Venkataravanappa,V., Jalali,S., KrishnaReddy,M. |
| Host | |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
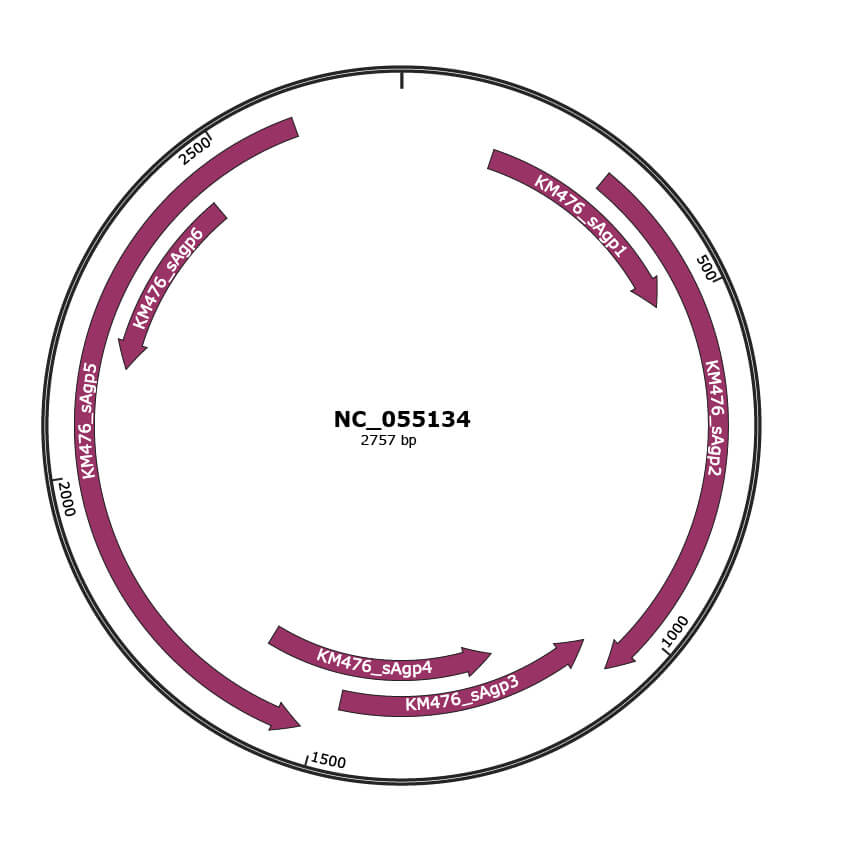
JBrowse
Genome
NC_055134
Gene Information
| NCBI Accession | YP_010084358.1 |
|---|---|
| Location | 143-499 |
| Protein Name | AV2 |
| Coding Region | ATGTGGGATCCATTGTTGAACGAGTTTCCCGAAACCGTTCACGGTTTTAGATGTATGTTAGCAGTTAAATATCTGCAGTTAGTAGAAAAGACTTATTCTCCTGACACATTAGGTTACGATTTAATTAGGGATTTAATTTCAGTTATTAGGGCTAGAAATTATGTCGAAGCGACCAGCAGATATCATCATTTCAACGCCCGCATCGAAGGTACGCCGACGTCTCAACTTCGACAGCCCCTATGGAGCTCGTGCAGTTGTCCCCATTGCCCGCGTCACAAAAGCAAAGGCCTGGACCAACAGGCCGATGAACAGAAAACCCAGAATGTACAGAATGTACAGGAGTCCCGACGTTCCTAG |
| Protein Sequence | MWDPLLNEFPETVHGFRCMLAVKYLQLVEKTYSPDTLGYDLIRDLISVIRARNYVEATSRYHHFNARIEGTPTSQLRQPLWSSCSCPHCPRHKSKGLDQQADEQKTQNVQNVQESRRS |
| NCBI Accession | YP_010084359.1 |
|---|---|
| Location | 303-1073 |
| Protein Name | AV1 |
| Coding Region | ATGTCGAAGCGACCAGCAGATATCATCATTTCAACGCCCGCATCGAAGGTACGCCGACGTCTCAACTTCGACAGCCCCTATGGAGCTCGTGCAGTTGTCCCCATTGCCCGCGTCACAAAAGCAAAGGCCTGGACCAACAGGCCGATGAACAGAAAACCCAGAATGTACAGAATGTACAGGAGTCCCGACGTTCCTAGGGGCTGTGAAGGCCCTTGTAAGGTGCAGTCTTTTGAATCTAGGCACGATGTCTCTCATATTGGCAAAGTCATGTGCGTTAGTGATGTTACTCGAGGAACTGGACTCACACATCGCGTGGGGAAGCGATTTTGTGTGAAATCTGTCTATGTGCTGGGGAAGATATGGATGGATGAAAACATCAAGACAAAAAACCATACTAACAGTGTCATGTTTTTTTTAGTTCGTGACCGTCGTCCTACAGGATCCCCCCAAGATTTTGGTGAAGTTTTTAACATGTTTGACAATGAACCGAGTACAGCAACGGTGAAGAACATGCATCGTGATCGTTATCAAGTGTTACGGAAGTGGCATGCGACTGTGACGGGAGGAACTTATGCATCTAAGGAGCAAGCATTAGTTAGGAAGTTTGTTAGGGTTAATAATTATGTTGTTTATAATCAACAAGAGGCAGGCAAGTATGAGAATCATACTGAAAATGCATTAATGTTGTATATGGCCTGTACTCACGCATCAAATCCTGTATACGCTACTTTGAAAATCCGGATCTATTTCTATGATTCGGTCACAAATTAA |
| Protein Sequence | MSKRPADIIISTPASKVRRRLNFDSPYGARAVVPIARVTKAKAWTNRPMNRKPRMYRMYRSPDVPRGCEGPCKVQSFESRHDVSHIGKVMCVSDVTRGTGLTHRVGKRFCVKSVYVLGKIWMDENIKTKNHTNSVMFFLVRDRRPTGSPQDFGEVFNMFDNEPSTATVKNMHRDRYQVLRKWHATVTGGTYASKEQALVRKFVRVNNYVVYNQQEAGKYENHTENALMLYMACTHASNPVYATLKIRIYFYDSVTN |
| NCBI Accession | YP_010084360.1 |
|---|---|
| Location | 1070-1474 |
| Protein Name | AC3 |
| Coding Region | ATGGATTCACGCACAGGGGAGTACATCACTGCGGATCGTACAGAGAGTGGCGTATATACCTGGGAAATTCGAAATCCCCTGTATTTCAAAATTTTGAAACACGACAGCAGGCCCTTCCTAACACAGCACGACATCATCAAGATTCAAATACAGTTCAACCACAACCTGCGGAAAGCCCTGGGACTTCACAAGTGTTTTCTAACTTTCAAAATCTGGACGGGCTTACGCCCTCAGATTGGGATTTTCTTGAGAGTCTTTAAAACCCAGGTCCTTAAGTACCTGGATATGTTAGGTGTCATAGGAATATATAATGTAATACGAGCAGTGTATCATGTGTTGGATAATGTATTGGAACAGACGATTGATGTATGGACTTCATATGATATAAAACTCGATATTTATTAA |
| Protein Sequence | MDSRTGEYITADRTESGVYTWEIRNPLYFKILKHDSRPFLTQHDIIKIQIQFNHNLRKALGLHKCFLTFKIWTGLRPQIGIFLRVFKTQVLKYLDMLGVIGIYNVIRAVYHVLDNVLEQTIDVWTSYDIKLDIY |
| NCBI Accession | YP_010084361.1 |
|---|---|
| Location | 1215-1619 |
| Protein Name | AC2 |
| Coding Region | ATGCAGTCTTCATCACACTCGAAGAACCACTCTATTCCGGTCGCGAAAACATCGCTCCCGCAGAAGAAGAAGAAGAGCATTCGCAGGAGGCGAGTTGATCTGCCGTGTGGGTGTTCGTATTACATATCGATCAACTGCCACGATCATGGATTCACGCACAGGGGAGTACATCACTGCGGATCGTACAGAGAGTGGCGTATATACCTGGGAAATTCGAAATCCCCTGTATTTCAAAATTTTGAAACACGACAGCAGGCCCTTCCTAACACAGCACGACATCATCAAGATTCAAATACAGTTCAACCACAACCTGCGGAAAGCCCTGGGACTTCACAAGTGTTTTCTAACTTTCAAAATCTGGACGGGCTTACGCCCTCAGATTGGGATTTTCTTGAGAGTCTTTAA |
| Protein Sequence | MQSSSHSKNHSIPVAKTSLPQKKKKSIRRRRVDLPCGCSYYISINCHDHGFTHRGVHHCGSYREWRIYLGNSKSPVFQNFETRQQALPNTARHHQDSNTVQPQPAESPGTSQVFSNFQNLDGLTPSDWDFLESL |
| NCBI Accession | YP_010084362.1 |
|---|---|
| Location | 1522-2607 |
| Protein Name | AC1 |
| Coding Region | ATGGCAGCCCCCAATCGGTTTAAAATAAATGCCAAAAATTATTTTCTCACTTATCCGAAGTGCTCTCTAACTAAAGAAGAAGCACTTTCCCAATTATTAAATATACATACACCCACTTCAAAAAAATTTATTAGAATTTGCAGAGAGCTTCACGAAGATGGGACTCCTCACTTGCATGTGCTCATCCAGTTCGAAGGAAAGTTCCAGTGCAAAAACAATAGATTCTTCGACCTCACATCCCCAACCAGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAGAGCTCAAGCGATGTCAAATCCTACATGGAAAAAGACGGAGACGTCATTGATCATGGAGTTTTCCAAGTCGATGGAAGATCAGCTAGAGGAGGTTGCCAATCTGCCAACGACGCATATGCCGAGGCAATCAATTCAGGGTCCAAAGCTCAGGCCCTCAATATACTGAAGGAGAAAGCTCCCAGAGACTTTGTACTTCAATTTCATAATTTAAATTCAAATTTAGATAGATATTTTCAGGATCCACCAGCTCTGTATATTTCTCCTTTTTCATCTTCTTCTTTCGACCAAGTTCCCGAAGAACTTGAAGAGTGGGCTTGCGAAAACATTGTCGATGCCGCTGCGCGGCCAATTAGACCCATAAGTATTGTTATTGAGGGTGATAGTAGTACTGGTAAAACAATGTGGGCCAGGTCATTGGGTTCACATAATTATCTGTGTGGTCATCTGGACCTTAGCCCAAAGATCTACAGCAATGATGCATGGTACAACGTCATTGATGACGTTGATCCCCACTATCTAAAGCACTTTAAAGAATTCATGGGGGCCCAGCGTGACTGGCAAAGCAACACCAAGTACGGAAAGCCAGTCATGATTAAAGGTGGAATTCCCACTATCTTCCTGTGCAATAAAGGTCCAAACAGCAGCTATAAGGAATATCTGGACGAAGAAAAAAATGCAGCACTGAAGCAGTGGGCAATCAAGAATGCAGTCTTCATCACACTCGAAGAACCACTCTATTCCGGTCGCGAAAACATCGCTCCCGCAGAAGAAGAAGAAGAGCATTCGCAGGAGGCGAGTTGA |
| Protein Sequence | MAAPNRFKINAKNYFLTYPKCSLTKEEALSQLLNIHTPTSKKFIRICRELHEDGTPHLHVLIQFEGKFQCKNNRFFDLTSPTRSAHFHPNIQGAKSSSDVKSYMEKDGDVIDHGVFQVDGRSARGGCQSANDAYAEAINSGSKAQALNILKEKAPRDFVLQFHNLNSNLDRYFQDPPALYISPFSSSSFDQVPEELEEWACENIVDAAARPIRPISIVIEGDSSTGKTMWARSLGSHNYLCGHLDLSPKIYSNDAWYNVIDDVDPHYLKHFKEFMGAQRDWQSNTKYGKPVMIKGGIPTIFLCNKGPNSSYKEYLDEEKNAALKQWAIKNAVFITLEEPLYSGRENIAPAEEEEEHSQEAS |
| NCBI Accession | YP_010084363.1 |
|---|---|
| Location | 2157-2450 |
| Protein Name | AC4 |
| Coding Region | ATGGGACTCCTCACTTGCATGTGCTCATCCAGTTCGAAGGAAAGTTCCAGTGCAAAAACAATAGATTCTTCGACCTCACATCCCCAACCAGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAGAGCTCAAGCGATGTCAAATCCTACATGGAAAAAGACGGAGACGTCATTGATCATGGAGTTTTCCAAGTCGATGGAAGATCAGCTAGAGGAGGTTGCCAATCTGCCAACGACGCATATGCCGAGGCAATCAATTCAGGGTCCAAAGCTCAGGCCCTCAATATACTGA |
| Protein Sequence | MGLLTCMCSSSSKESSSAKTIDSSTSHPQPGQHISIRTFRELRAQAMSNPTWKKTETSLIMEFSKSMEDQLEEVANLPTTHMPRQSIQGPKLRPSIY |
References More References in PubMed
| 1 |
Naganur P, et al. Plants (Basel). 2023 Jan 20;12(3):490. doi: 10.3390/plants12030490. PMID: 36771575 |
|---|---|
| 2 |
Venkataravanappa V, et al. 3 Biotech. 2020 Jun;10(6):282. doi: 10.1007/s13205-020-02245-x. Epub 2020 Jun 2. PMID: 32550101 |
| 3 |
A New Begomovirus Species Causing Tomato Leaf Curl Disease in Varanasi, India. Chakraborty S, et al. Plant Dis. 2003 Mar;87(3):313. doi: 10.1094/PDIS.2003.87.3.313A. PMID: 30812767 |
| 4 |
Prasad KSUD, et al. Virusdisease. 2024 Sep;35(3):484-495. doi: 10.1007/s13337-024-00891-w. Epub 2024 Sep 16. PMID: 39464737 |
| 5 |
Banks GK, et al. Plant Dis. 2001 Feb;85(2):231. doi: 10.1094/PDIS.2001.85.2.231C. PMID: 30831961 |
| 6 |
Kiran GVNSM, et al. 3 Biotech. 2021 Dec;11(12):500. doi: 10.1007/s13205-021-02975-6. Epub 2021 Nov 17. PMID: 34881163 |
| 7 |
Rathore S, et al. Plant Dis. 2014 Mar;98(3):428. doi: 10.1094/PDIS-07-13-0719-PDN. PMID: 30708408 |
| 8 |
Devaraj, et al. 3 Biotech. 2025 Aug;15(8):260. doi: 10.1007/s13205-025-04434-y. Epub 2025 Jul 19. PMID: 40693173 |
| 9 |
Möller SR, et al. Mol Plant Pathol. 2026 Mar;27(3):e70202. doi: 10.1111/mpp.70202. PMID: 41761047 |
| 10 |
Nayaka SN, et al. Virusdisease. 2023 Sep;34(3):421-430. doi: 10.1007/s13337-023-00837-8. Epub 2023 Sep 8. PMID: 37780909 |