Tomato leaf curl Karnataka virus 2
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_004787115.1 |
| Isolate | India: Sonipet, Hariyana |
| Release date | 2021/6/1 |
| Submitter | Swarnalatha,S., Venkataravanappa,V., Jalali,S., Krishnareddy,M. |
| Host | |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
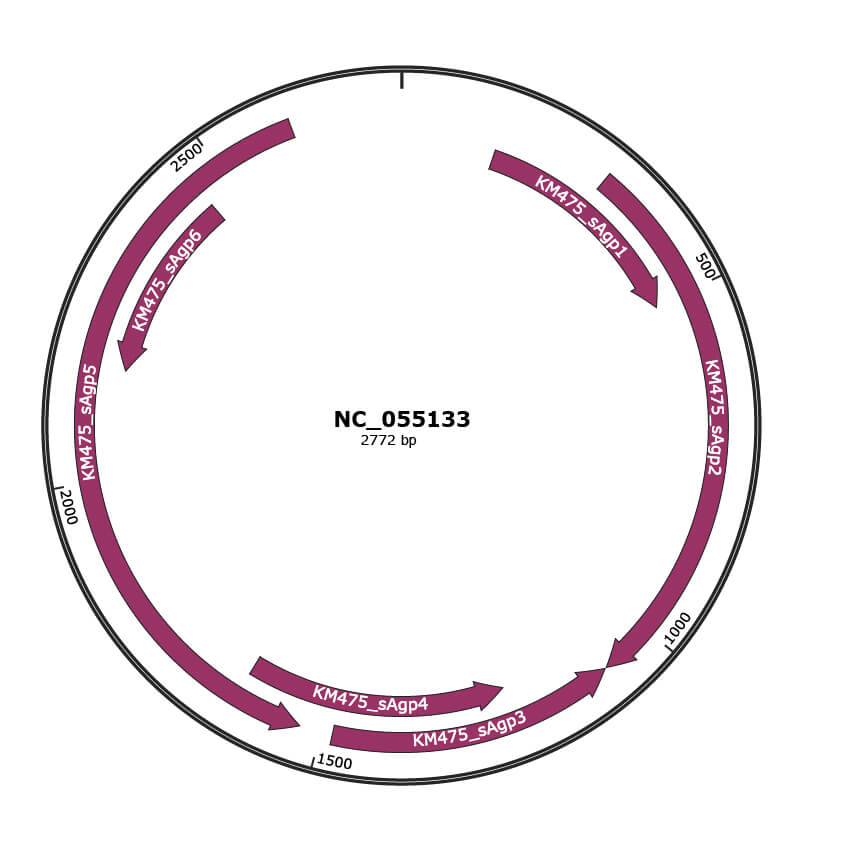
JBrowse
Genome
NC_055133
Gene Information
| NCBI Accession | YP_010084352.1 |
|---|---|
| Location | 146-502 |
| Protein Name | AV2 |
| Coding Region | ATGTGGGATCCATTATTGCACGAATTTCCAGAAAGCGTTCATGGTCTAAGGTGCATGCTAGCTGTAAAATATCTGCAAGAGATAGAAAAGAACTATTCGCCAGACACAGTCGGATACGATCTTGTCCGAGATCTCATTCTTGTTCTGCGAGCAAAGAACTATGGCGAAGCGACCAGCAGATATCATCATTTCAACGCCCGCATCGAAAGTACGCCGACGTCTCAACTTCGACAGCCCCTATGGAGCTCGTGCAGTTGTCCCCATTGCCCGCGTCACAAAAGCAAAGGCCTGGACAAACAGGCCGATGAACAGAAAACCCAGAATGTACAGAATGTACAGAAGTCCCGACGTGCCTAG |
| Protein Sequence | MWDPLLHEFPESVHGLRCMLAVKYLQEIEKNYSPDTVGYDLVRDLILVLRAKNYGEATSRYHHFNARIESTPTSQLRQPLWSSCSCPHCPRHKSKGLDKQADEQKTQNVQNVQKSRRA |
| NCBI Accession | YP_010084353.1 |
|---|---|
| Location | 306-1076 |
| Protein Name | AV1 |
| Coding Region | ATGGCGAAGCGACCAGCAGATATCATCATTTCAACGCCCGCATCGAAAGTACGCCGACGTCTCAACTTCGACAGCCCCTATGGAGCTCGTGCAGTTGTCCCCATTGCCCGCGTCACAAAAGCAAAGGCCTGGACAAACAGGCCGATGAACAGAAAACCCAGAATGTACAGAATGTACAGAAGTCCCGACGTGCCTAGGGGATGTGAAGGCCCTTGCAAGGTGCAGTCCTTTGAATCAAGGCACGATGTCTCTCATATTGGGAAAGTGATGTGCGTTAGTGATGTTACCCGAGGAACTGGACTCACACATCGCGTAGGAAAGCGATTTTGTGTGAAATCTGTCTATGTGCTGGGGAAGATATGGATGGATGAAAACATCAAGACGAAGAACCATACTAACAGTGTTATGTTTTTTTTGGTACGTGACCGTCGTCCTACAGGATCCCCCCAAGATTTTGGGGAAGTTTTTAACATGTTTGACAATGAACCGAGCACAGCAACGGTGAAGAACATGCATCGTGATCGTTATCAAGTGTTACGGAAGTGGCATGCGACTGTGACGGGAGGAACATATGCATGTAGGGAGCAAGCATTAGTTAGGAAGTTTGTTAGGGTTAATAATTATGTAGTTTATAATCAACAAGAGGCCGGCAAGTATGAGAATCATACTGAAAACGCATTAATGTTGTATATGGCCTGTACTCATGCCTCTAATCCTGTGTATGCTACTTTGAAAGTTAGGAGTTACTTCTACGATTCTGTAACAAATTAA |
| Protein Sequence | MAKRPADIIISTPASKVRRRLNFDSPYGARAVVPIARVTKAKAWTNRPMNRKPRMYRMYRSPDVPRGCEGPCKVQSFESRHDVSHIGKVMCVSDVTRGTGLTHRVGKRFCVKSVYVLGKIWMDENIKTKNHTNSVMFFLVRDRRPTGSPQDFGEVFNMFDNEPSTATVKNMHRDRYQVLRKWHATVTGGTYACREQALVRKFVRVNNYVVYNQQEAGKYENHTENALMLYMACTHASNPVYATLKVRSYFYDSVTN |
| NCBI Accession | YP_010084354.1 |
|---|---|
| Location | 1079-1483 |
| Protein Name | AC3 |
| Coding Region | ATGGATTCACGCACAGGGGAACCCATCACTGCAGCTCAAGCACAGAATGGCGTGTTTATCTGGGAGATTCAAAATCCCCTGTATTTCAAGATAACAGAACACCACAACAGGCCATTCCTAATGAAGGAAGACATCATCACCGTTCAGATACAGTTCAATCACAACCTGAGGAAAGCGTTGGGGATACACAAATGTTTCCTAGCCTTCCGAATCTGGATGACCTCACAGCCTCCGACTGGTCGTTTCTTAAGGGTCTTTAAGACTCAAGTTCTTAAGTATTTAAATAACTTAGGAGTTATCAGTATTAACACTGTAATTAAATCAGTTGATCATGTATTATGGAATGTATTACATCACGTTGTATATGTAGACCAATCATATTCAATAAAATTCAATATTTATTAA |
| Protein Sequence | MDSRTGEPITAAQAQNGVFIWEIQNPLYFKITEHHNRPFLMKEDIITVQIQFNHNLRKALGIHKCFLAFRIWMTSQPPTGRFLRVFKTQVLKYLNNLGVISINTVIKSVDHVLWNVLHHVVYVDQSYSIKFNIY |
| NCBI Accession | YP_010084355.1 |
|---|---|
| Location | 1224-1628 |
| Protein Name | AC2 |
| Coding Region | ATGCGATCTTCGTCACCCTCGAAGGCCCATTGTACTCAGGTTCCCATCAAAGTGCAGCACAGGCTAGCCAAGAAGGGGACCAGACGTCGACGTGTTGATCTACCGTGCGGCTGTTCATACTTCATTGCATTAGCCTGCCACAACCATGGATTCACGCACAGGGGAACCCATCACTGCAGCTCAAGCACAGAATGGCGTGTTTATCTGGGAGATTCAAAATCCCCTGTATTTCAAGATAACAGAACACCACAACAGGCCATTCCTAATGAAGGAAGACATCATCACCGTTCAGATACAGTTCAATCACAACCTGAGGAAAGCGTTGGGGATACACAAATGTTTCCTAGCCTTCCGAATCTGGATGACCTCACAGCCTCCGACTGGTCGTTTCTTAAGGGTCTTTAA |
| Protein Sequence | MRSSSPSKAHCTQVPIKVQHRLAKKGTRRRRVDLPCGCSYFIALACHNHGFTHRGTHHCSSSTEWRVYLGDSKSPVFQDNRTPQQAIPNEGRHHHRSDTVQSQPEESVGDTQMFPSLPNLDDLTASDWSFLKGL |
| NCBI Accession | YP_010084356.1 |
|---|---|
| Location | 1531-2616 |
| Protein Name | AC1 |
| Coding Region | ATGGCTCCGCCAAGTCGTTTTAGAATAAATGCGAAAAACTATTTTCTCACATATCTCAAGTGCTCTCTAACTAAAGAAGAGGCACTTTCCCAATTATTAAATCTCCAAACACCCACTTCCAAAAAATTTATTAGAATTTGCAGAGAGCTTCACGAAGATGGGACTCCTCACTTGCATGTGCTCATCCAGTTCGAAGGAAAGTTCCAGTGCAAAAACAATAGATTCTTCGACCTTACATCCCCAACCAGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAGAGCTCAAGCGATGTCAAATCCTACATGGAAAAAGACGGAGACGTCCTTGATCATGGAGTTTTCCAAGTCGATGGACGATCAGCTAGAGGAGGTTGCCAATCTGCCAACGACGCATATGCCGAGGCAATCAACTCAGGGTCCAAAGCTCAGGCCCTCAATATACTGAAGGAGAAAGCTCCCAGAGACTTTCTATTTCAATTTCATAATTTAAATTCTAATTTAGATAGGTTTTTTACACCTCCAATGGAGGTTTATGTTTCTCCTTTTTTATCTTCATCTTTTGATCAAGTTCCCGAAGAACTTGAGGAGTGGGCTGCAGAGAATGTGGTGAGATCCGCTGCGCGGCCTTTGAGACCCGTAAGTATAGTCATTGAGGGTGATAGTAGGACAGGGAAGACTATGTGGGCCAGATCACTGGGACCACATAATTATTTGTGTGGTCATTTAGACCTTAGTCCAAAGGTCTACAATAATGATGCCTGGTATAACGTCATTGATGACGTAGACCCCCACTATCTAAAGCACTTTAAAGAATTCATGGGGGCCCAAAGGGACTGGCAATCAAATACAAAGTACGGGAAGCCAGTTCAAATTAAAGGCGGAATTCCCACTATCTTCCTCTGCAATCCAGGACCAAATTCCAGCTATAAAGAGTTCTTGGATGAAGATAAGAATTCTGCACTAAAAAACTGGGCATTAAAAAATGCGATCTTCGTCACCCTCGAAGGCCCATTGTACTCAGGTTCCCATCAAAGTGCAGCACAGGCTAGCCAAGAAGGGGACCAGACGTCGACGTGTTGA |
| Protein Sequence | MAPPSRFRINAKNYFLTYLKCSLTKEEALSQLLNLQTPTSKKFIRICRELHEDGTPHLHVLIQFEGKFQCKNNRFFDLTSPTRSAHFHPNIQGAKSSSDVKSYMEKDGDVLDHGVFQVDGRSARGGCQSANDAYAEAINSGSKAQALNILKEKAPRDFLFQFHNLNSNLDRFFTPPMEVYVSPFLSSSFDQVPEELEEWAAENVVRSAARPLRPVSIVIEGDSRTGKTMWARSLGPHNYLCGHLDLSPKVYNNDAWYNVIDDVDPHYLKHFKEFMGAQRDWQSNTKYGKPVQIKGGIPTIFLCNPGPNSSYKEFLDEDKNSALKNWALKNAIFVTLEGPLYSGSHQSAAQASQEGDQTSTC |
| NCBI Accession | YP_010084357.1 |
|---|---|
| Location | 2166-2459 |
| Protein Name | AC4 |
| Coding Region | ATGGGACTCCTCACTTGCATGTGCTCATCCAGTTCGAAGGAAAGTTCCAGTGCAAAAACAATAGATTCTTCGACCTTACATCCCCAACCAGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAGAGCTCAAGCGATGTCAAATCCTACATGGAAAAAGACGGAGACGTCCTTGATCATGGAGTTTTCCAAGTCGATGGACGATCAGCTAGAGGAGGTTGCCAATCTGCCAACGACGCATATGCCGAGGCAATCAACTCAGGGTCCAAAGCTCAGGCCCTCAATATACTGA |
| Protein Sequence | MGLLTCMCSSSSKESSSAKTIDSSTLHPQPGQHISIRTFRELRAQAMSNPTWKKTETSLIMEFSKSMDDQLEEVANLPTTHMPRQSTQGPKLRPSIY |
References More References in PubMed
| 1 |
Tomato leaf curl Karnataka virus from Bangalore, India, Appears to be a Recombinant Begomovirus. Chatchawankanphanich O, et al. Phytopathology. 2002 Jun;92(6):637-45. doi: 10.1094/PHYTO.2002.92.6.637. PMID: 18944261 |
|---|---|
| 2 |
Venkataravanappa V, et al. 3 Biotech. 2020 Jun;10(6):282. doi: 10.1007/s13205-020-02245-x. Epub 2020 Jun 2. PMID: 32550101 |
| 3 |
A New Begomovirus Species Causing Tomato Leaf Curl Disease in Varanasi, India. Chakraborty S, et al. Plant Dis. 2003 Mar;87(3):313. doi: 10.1094/PDIS.2003.87.3.313A. PMID: 30812767 |
| 4 |
Chakraborty S, et al. Phytopathology. 2003 Dec;93(12):1485-95. doi: 10.1094/PHYTO.2003.93.12.1485. PMID: 18943612 |
| 5 |
Prasad KSUD, et al. Virusdisease. 2024 Sep;35(3):484-495. doi: 10.1007/s13337-024-00891-w. Epub 2024 Sep 16. PMID: 39464737 |
| 6 |
Venkataravanappa V, et al. Iran J Biotechnol. 2019 Jan 11;17(1):e2134. doi: 10.21859/ijb.2134. eCollection 2019 Jan. PMID: 31457044 |
| 7 |
Rathore S, et al. Plant Dis. 2014 Mar;98(3):428. doi: 10.1094/PDIS-07-13-0719-PDN. PMID: 30708408 |
| 8 |
Banks GK, et al. Plant Dis. 2001 Feb;85(2):231. doi: 10.1094/PDIS.2001.85.2.231C. PMID: 30831961 |
| 9 |
Venkataravanappa V, et al. Microb Pathog. 2023 Jul;180:106127. doi: 10.1016/j.micpath.2023.106127. Epub 2023 Apr 28. PMID: 37119939 |
| 10 |
Venkataravanappa V, et al. Virusdisease. 2022 Jun;33(2):194-207. doi: 10.1007/s13337-022-00772-0. Epub 2022 Jul 1. PMID: 35991698 |