Tomato leaf curl Joydebpur virus
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_000864425.1 |
| Isolate | Bangladesh:Gazipur, Joydebpur |
| Release date | 2015/2/13 |
| Submitter | Maruthi,M.N., Rekha,A.R., Alam,S.N., Kader,K.A., Cork,A., Colvin,J. |
| Host | |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
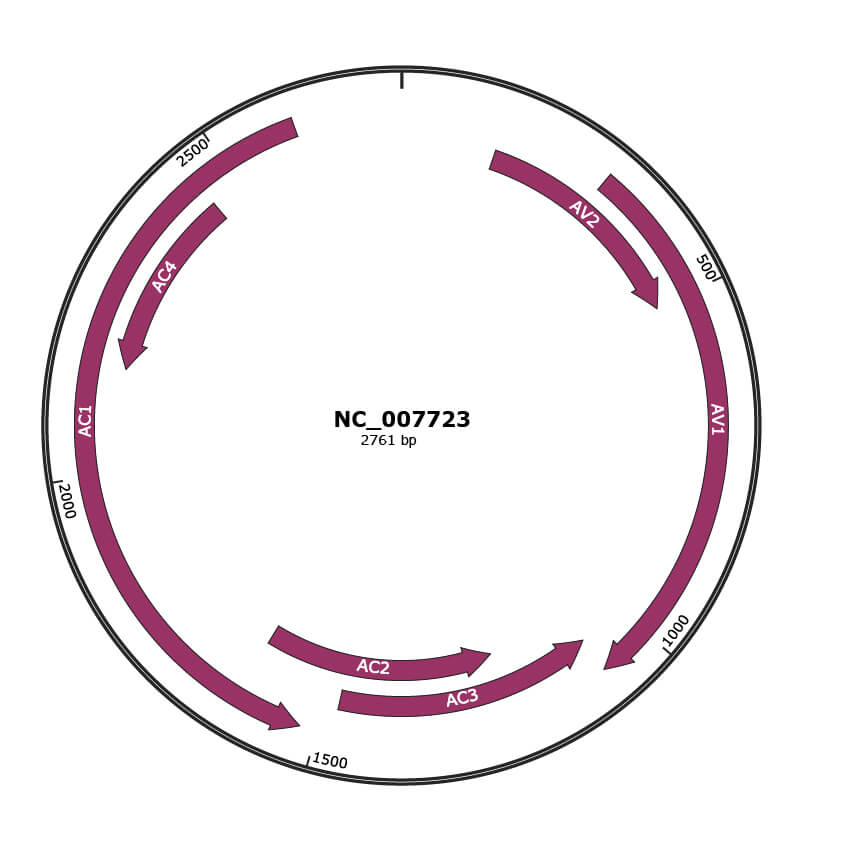
JBrowse
Genome
NC_007723
Gene Information
| NCBI Accession | YP_459901.1 |
|---|---|
| Location | 146-502 |
| Gene Name | AV2 |
| Protein Name | AV2 protein |
| Coding Region | ATGTGGGATCCGTTAGTAAACGACTTTCCTGAAACCGTTCACGGTTTTAGGTGTATGCTAGCCGTTAAATATCTGCAGCTAGTAGAAAAGACGTATTCTCCAGATACTCTCGGGTACGATTTAATCCGGGATTTAATTTCAGTTATCAGGGCCAGAAATTATGGCGAAGCGACCGGCAGATATAATCATTTCCACGCCCGCCTCGAAGGTACGCCGCCGTCTCAACTTCGACAGCCCATATTCCAGCCGTGCTGCTGCCCCCATTGTCCGCGTCACCAAAGCAAAAGCATGGGCGAACAGGCCCATGAACAGGAAGCCCAGGATGTACAGGATGTTCAGAAGCCCAGATGTTCCTAG |
| Protein Sequence | MWDPLVNDFPETVHGFRCMLAVKYLQLVEKTYSPDTLGYDLIRDLISVIRARNYGEATGRYNHFHARLEGTPPSQLRQPIFQPCCCPHCPRHQSKSMGEQAHEQEAQDVQDVQKPRCS |
| NCBI Accession | YP_459902.1 |
|---|---|
| Location | 306-1076 |
| Gene Name | AV1 |
| Protein Name | coat protein |
| Coding Region | ATGGCGAAGCGACCGGCAGATATAATCATTTCCACGCCCGCCTCGAAGGTACGCCGCCGTCTCAACTTCGACAGCCCATATTCCAGCCGTGCTGCTGCCCCCATTGTCCGCGTCACCAAAGCAAAAGCATGGGCGAACAGGCCCATGAACAGGAAGCCCAGGATGTACAGGATGTTCAGAAGCCCAGATGTTCCTAGAGGCTGTGAAGGCCCATGTAAGGTCCAGTCCTTTGAGTCTAGACATGATGTCCAACACATAGGTAAAGTCATGTGTGTTAGTGATGTTACTCGTGGAACTGGGCTGACCCATCGAGTTGGTAAAAGGTTTTGTGTTAAGTCCGTCTATGTACTGGGCAAGATTTGGATGGATGAGAACATCAAGACTAAGAATCACACGAATAGTGTTATGTTTTTCCTTGTTAGAGATCGTAGGCCCGTAGATAAGCCCCAAGACTTTGGTGAGGTGTTTAACATGTTTGATAATGAGCCCAGTACGGCTACCGTGAAGAACGTGCATCGTGATAGGTATCAGGTGCTCAGGAAGTGGCATGCAACTGTGACTGGTGGTCAATATGCATCGAAGGAGCAAGCTCTCGTGAAGAAGTTTATTAGGGTTAATAATTATGTTGTGTATAACCAGCAAGAGGCTGGCAAGTATGAAAACCATACTGAGAATGCATTGATGTTGTACATGGCGTGTACTCATGCCTCTAACCCTGTGTATGCTACTTTGAAGATACGGATCTATTTCTATGATTCCGTAACAAATTAA |
| Protein Sequence | MAKRPADIIISTPASKVRRRLNFDSPYSSRAAAPIVRVTKAKAWANRPMNRKPRMYRMFRSPDVPRGCEGPCKVQSFESRHDVQHIGKVMCVSDVTRGTGLTHRVGKRFCVKSVYVLGKIWMDENIKTKNHTNSVMFFLVRDRRPVDKPQDFGEVFNMFDNEPSTATVKNVHRDRYQVLRKWHATVTGGQYASKEQALVKKFIRVNNYVVYNQQEAGKYENHTENALMLYMACTHASNPVYATLKIRIYFYDSVTN |
| NCBI Accession | YP_459903.1 |
|---|---|
| Location | 1073-1477 |
| Gene Name | AC3 |
| Protein Name | replication enhancer |
| Coding Region | ATGGATTCACGCACAGGGGAACCCATCACTGCAGCTCAGGCCGAGAATGGCGTGTTTATCTGGGAGATAACAAATCCCCTCTATTTCAAGATAACCGAGCACCACCACCGGCCATTCCTAATGAACCACGACATCATCACAGTCCAAGTACAGTTCAATCACAACCTGAGGAAAGCGTTGGGGATACACAAATGTTTTCTAACCTTCCGAATCTGGACGACCTTACAGCCTCGGACTGGTCATTTCTTAAGGGTCTTTAGGACCCAAGTTTTGCAGTTTTTAAATAATTTTGCTGTAATTAGTATAAACAATGTAATTAGAGCAGTCGATCATGTATTATGGAATGTATTAGCGCAAACAATTTATGTACAAAGTTCAAGCATAATAAAATTCAATATTTATTAA |
| Protein Sequence | MDSRTGEPITAAQAENGVFIWEITNPLYFKITEHHHRPFLMNHDIITVQVQFNHNLRKALGIHKCFLTFRIWTTLQPRTGHFLRVFRTQVLQFLNNFAVISINNVIRAVDHVLWNVLAQTIYVQSSSIIKFNIY |
| NCBI Accession | YP_459904.1 |
|---|---|
| Location | 1218-1622 |
| Gene Name | AC2 |
| Protein Name | transcriptional activator protein (TrAP) |
| Coding Region | ATGCGACCTTCATCACCCTCGAAGGCCCACTCTACTCAGGTTCCAATCAAAGTCCAGCACAGGCTAGCCAAGAGGGGGACCAGGCGTCGTCGCGTTGACCTGGACTGCGGCTGTTCATACTTCATCGCATTACGCTGCCACGACTATGGATTCACGCACAGGGGAACCCATCACTGCAGCTCAGGCCGAGAATGGCGTGTTTATCTGGGAGATAACAAATCCCCTCTATTTCAAGATAACCGAGCACCACCACCGGCCATTCCTAATGAACCACGACATCATCACAGTCCAAGTACAGTTCAATCACAACCTGAGGAAAGCGTTGGGGATACACAAATGTTTTCTAACCTTCCGAATCTGGACGACCTTACAGCCTCGGACTGGTCATTTCTTAAGGGTCTTTAG |
| Protein Sequence | MRPSSPSKAHSTQVPIKVQHRLAKRGTRRRRVDLDCGCSYFIALRCHDYGFTHRGTHHCSSGREWRVYLGDNKSPLFQDNRAPPPAIPNEPRHHHSPSTVQSQPEESVGDTQMFSNLPNLDDLTASDWSFLKGL |
| NCBI Accession | YP_459905.1 |
|---|---|
| Location | 1525-2610 |
| Gene Name | AC1 |
| Protein Name | replication protein |
| Coding Region | ATGGCTCCTCCAAATAAATTCAGAATAAATGCCAAAAATTATTTCCTCACATATCCCAAGTGCTCTCTTACTAAAGAAGAGGCACTTTCCCAATTACAACATTTAGAAACTCCAGTAAATAAATTATTTATTAGGGTTTGCAGAGAACTACACGAAAATGGGGAACCTCATCTACACGTGCTCCTCCAATTTGAGGGAAAATTCCAATGCAAAAATCACCGATTCTTCGACCTCGTTTCCCCATCCCGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAGAGCTCTACAGATGTCAAAGCCTACGTGGAGAAAGACGGAGACTTCATTGATTTTGGAGTTTTCCAGATCGATGGCAGATCAGCTAGAGGAGGTCAGCAATCTGCCAACGACGCATATGCCGAGGCAATCAATTCATGGTCCAAAGCTGCGGCCCTCAATATTTTAAGGGAGAAAGCTCCCAAAGATTATGTTTTACAGTTTCATAATTTAAATGCTAATTTAGATAGGATTTTTACACCTCCACCGGAGGTTTATGTTTCTCCTTTTTCTTCCTCTTCTTTTGATCAAGTTCCCGAGGAACTTGAAGAATGGGCTACCGAGAATGTGGTGTCTGCCGCTGCGCGGCCATTACGACCCGTAAGTGTAGTCATACAGGGTGATAGTCGAACGGGCAAGACGGTGTGGGCGAGGTCACTAGGTCCACACAATTATTTGTGTGGTCATCTCGACCTTAGCCCAAAGGTGTACAGTAATGACGCCTGGTACAACGTCATTGATGACGTTGATCCTCATTATCTAAAACACTTTAAAGAATTCATGGGGGCCCAAAGGGACTGGCAATCAAATACAAAATACGGAAAGCCAGTTCAAATTAAAGGTGGAATTCCCGCTATCTTCCTCTGCAATCCAGGACCCAATTCCAGCTATAAAGAGTTCTTGGATGAAGAGAAAAATAGCGCACTCAAAAATTGGGCTTTGAAGAATGCGACCTTCATCACCCTCGAAGGCCCACTCTACTCAGGTTCCAATCAAAGTCCAGCACAGGCTAGCCAAGAGGGGGACCAGGCGTCGTCGCGTTGA |
| Protein Sequence | MAPPNKFRINAKNYFLTYPKCSLTKEEALSQLQHLETPVNKLFIRVCRELHENGEPHLHVLLQFEGKFQCKNHRFFDLVSPSRSAHFHPNIQGAKSSTDVKAYVEKDGDFIDFGVFQIDGRSARGGQQSANDAYAEAINSWSKAAALNILREKAPKDYVLQFHNLNANLDRIFTPPPEVYVSPFSSSSFDQVPEELEEWATENVVSAAARPLRPVSVVIQGDSRTGKTVWARSLGPHNYLCGHLDLSPKVYSNDAWYNVIDDVDPHYLKHFKEFMGAQRDWQSNTKYGKPVQIKGGIPAIFLCNPGPNSSYKEFLDEEKNSALKNWALKNATFITLEGPLYSGSNQSPAQASQEGDQASSR |
| NCBI Accession | YP_459906.1 |
|---|---|
| Location | 2160-2453 |
| Gene Name | AC4 |
| Protein Name | AC4 protein |
| Coding Region | ATGGGGAACCTCATCTACACGTGCTCCTCCAATTTGAGGGAAAATTCCAATGCAAAAATCACCGATTCTTCGACCTCGTTTCCCCATCCCGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAGAGCTCTACAGATGTCAAAGCCTACGTGGAGAAAGACGGAGACTTCATTGATTTTGGAGTTTTCCAGATCGATGGCAGATCAGCTAGAGGAGGTCAGCAATCTGCCAACGACGCATATGCCGAGGCAATCAATTCATGGTCCAAAGCTGCGGCCCTCAATATTTTAA |
| Protein Sequence | MGNLIYTCSSNLRENSNAKITDSSTSFPHPGQHISIRTFRELRALQMSKPTWRKTETSLILEFSRSMADQLEEVSNLPTTHMPRQSIHGPKLRPSIF |
References More References in PubMed
| 1 |
Vo TTB, et al. Microorganisms. 2023 Dec 1;11(12):2907. doi: 10.3390/microorganisms11122907. PMID: 38138051 |
|---|---|
| 2 |
Bottle gourd IC-0262269, a super-susceptible genotype to tomato leaf curl Palampur virus. Nayaka SN, et al. 3 Biotech. 2024 Jan;14(1):8. doi: 10.1007/s13205-023-03838-y. Epub 2023 Dec 8. PMID: 38074288 |
| 3 |
Tiwari N, et al. Arch Virol. 2013 Jan;158(1):1-10. doi: 10.1007/s00705-012-1440-6. Epub 2012 Aug 24. PMID: 22918555 |
| 4 |
Silencing of tomato CTR1 provides enhanced tolerance against Tomato leaf curl virus infection. Chandan RK, et al. Plant Signal Behav. 2019;14(3):e1565595. doi: 10.1080/15592324.2019.1565595. Epub 2019 Jan 19. PMID: 30661468 |
| 5 |
Krishnan N, et al. J Virol Methods. 2022 Apr;302:114474. doi: 10.1016/j.jviromet.2022.114474. Epub 2022 Jan 22. PMID: 35077721 |
| 6 |
First Report of Tomato leaf curl New Delhi virus Infecting Tomato in Bangladesh. Maruthi MN, et al. Plant Dis. 2005 Sep;89(9):1011. doi: 10.1094/PD-89-1011C. PMID: 30786642 |
| 7 |
A New Begomovirus Species Causing Tomato Leaf Curl Disease in Patna, India. Kumari P, et al. Plant Dis. 2009 May;93(5):545. doi: 10.1094/PDIS-93-5-0545B. PMID: 30764166 |
| 8 |
Kumari P, et al. Virus Res. 2010 Sep;152(1-2):19-29. doi: 10.1016/j.virusres.2010.05.015. Epub 2010 Jun 9. PMID: 20540978 |
| 9 |
Nayaka SN, et al. Virusdisease. 2023 Sep;34(3):421-430. doi: 10.1007/s13337-023-00837-8. Epub 2023 Sep 8. PMID: 37780909 |
| 10 |
Maruthi MN, et al. Phytopathology. 2005 Dec;95(12):1472-81. doi: 10.1094/PHYTO-95-1472. PMID: 18943559 |