Tomato leaf curl Japan virus
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_004786335.1 |
| Isolate | Japan: Miyazaki |
| Release date | 2021/6/1 |
| Submitter | Ueda,S., Onuki,M., Hanada,K., Takanami,Y. |
| Host | |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
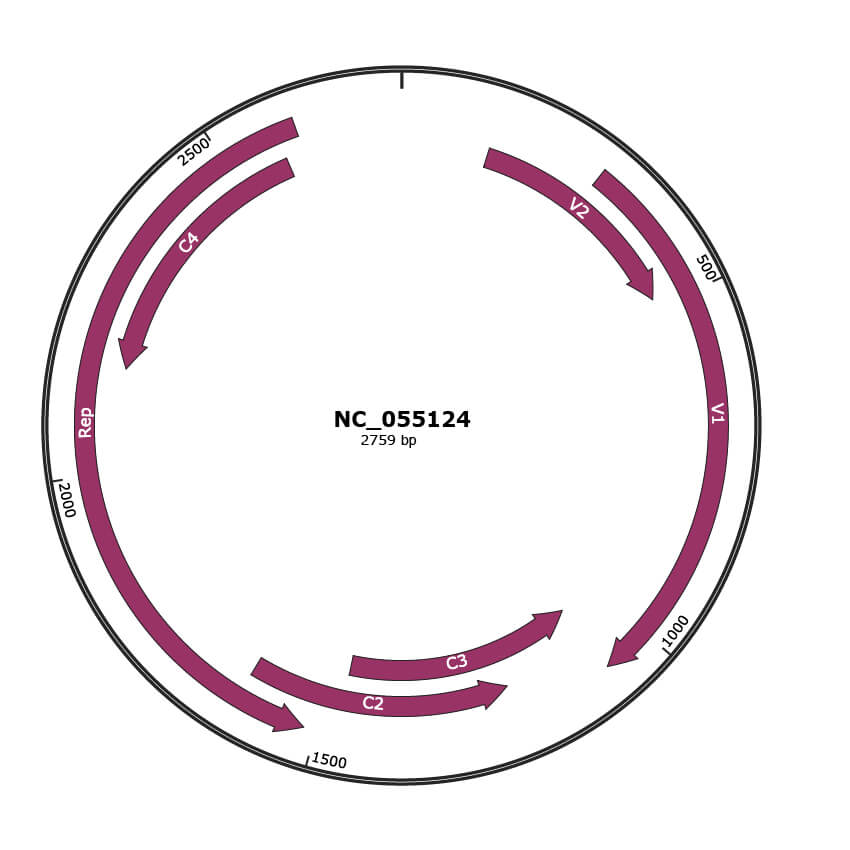
JBrowse
Genome
NC_055124
Gene Information
| NCBI Accession | YP_010084308.1 |
|---|---|
| Location | 136-486 |
| Gene Name | V2 |
| Protein Name | V2 |
| Coding Region | ATGTGGGATCCTTTACTTAACGATTTCCCTGAAACCGTTCACGGTTTCAGGTGTATGCTAGCGGTTAAGTACCTTCTACTCGTTGAAGCTACCTACTCTCCGGATACTATAGGTTACGATCTGATTCGTGATCTTGTGGGTGTTGTTCGTGCCAAGAACTATGTCGAAGCGTCCTGCAGATATAGGAATTTTCACTCCCGTCTCCAGAGCACGACGCCGTCTGAACTTCGACAGCCCGTTGCTCAGCCGTGCGAGTGCCCCCATTGCCCTCGTCACAAACAAAAAGAGATCATGGGCTCAGAGGCCCATGTATCGGAAGCCCAAGATGTACAGAATGTACAGAAGTCCTGA |
| Protein Sequence | MWDPLLNDFPETVHGFRCMLAVKYLLLVEATYSPDTIGYDLIRDLVGVVRAKNYVEASCRYRNFHSRLQSTTPSELRQPVAQPCECPHCPRHKQKEIMGSEAHVSEAQDVQNVQKS |
| NCBI Accession | YP_010084309.1 |
|---|---|
| Location | 296-1069 |
| Gene Name | V1 |
| Protein Name | coat protein |
| Coding Region | ATGTCGAAGCGTCCTGCAGATATAGGAATTTTCACTCCCGTCTCCAGAGCACGACGCCGTCTGAACTTCGACAGCCCGTTGCTCAGCCGTGCGAGTGCCCCCATTGCCCTCGTCACAAACAAAAAGAGATCATGGGCTCAGAGGCCCATGTATCGGAAGCCCAAGATGTACAGAATGTACAGAAGTCCTGATATCCCCAAGGGGTGTGAAGGCCCGTGTAAGGTCCAGTCTTTCGAGAAGAAAGACGATGTTGGTCATTCTGGTAAAATGCTCTGTATTTCTGATATCACCCGTGGCAACGGACTTAATCACCGTGTTGGGAAGAGATTTGGTATAAAATCTGTTTACATTATGGGAAAGATCTGGATGGATGAGAATATTAAGCTTAAAAATCACACTAACAACGTTTTATTTTGGTTAGTTAGAGATAGACGCCCTGTTACTACCCCTTATGCATTCCAAGAGGCATTTAATATGTTTGAGATGGAACCCAGTACGGCTACTATCAAGCAGGATTTGAGAGATCGGTTGCAGGTCTTACACAAGTTCAGTGCTACTGTCACTGGTGGACAGTATGCGTCCAGGGAGCAAGCTCTTATCAAGAGATTCTGGAAGTTGAATCATCATGTCACTTACAATCATCAAGAAGCTGCTAAATATGAGAATCACACTGAAAATGCTTTGTTATTGTATATGGCTTGCACTCATGCCAGTAATCCAGTGTATGCAACATTAAAAATTAGAGTGTATTTCTATGATTCAGAACAGAATTAA |
| Protein Sequence | MSKRPADIGIFTPVSRARRRLNFDSPLLSRASAPIALVTNKKRSWAQRPMYRKPKMYRMYRSPDIPKGCEGPCKVQSFEKKDDVGHSGKMLCISDITRGNGLNHRVGKRFGIKSVYIMGKIWMDENIKLKNHTNNVLFWLVRDRRPVTTPYAFQEAFNMFEMEPSTATIKQDLRDRLQVLHKFSATVTGGQYASREQALIKRFWKLNHHVTYNHQEAAKYENHTENALLLYMACTHASNPVYATLKIRVYFYDSEQN |
| NCBI Accession | YP_010084310.1 |
|---|---|
| Location | 1066-1470 |
| Gene Name | C3 |
| Protein Name | C3 |
| Coding Region | ATGGATTCACGCACAGGGGTCACCATCACTGCAGCTCAGGCACAGAGTGGCGCGTATACTTGGACAGTGCCAAATCCCCTATATTTCAAAATCACGAATCACGCCCAACGGCCATTCAACATGAATCAGGACATAATCACAGTACAGATACAATTCAACCACAACCTCCGATCTCAGCTGGATTTACACAAGTGCTTCCTGACTTTCAAGATTTGGACTCTCTCACATCATCAGAACTCGCATTTCTTGAATTTATTTAGGGAACATGTGTTGATGTATTTAGATAACTTGGGTGTTATTTCAATTAACAATGTAATAAGGGCAGTTGAATATGTATTGTATGATGTATTAGAAGGAACGGAGTTTGTTCAACAATTTACTGATATAAAATTCAAACTTTATTAA |
| Protein Sequence | MDSRTGVTITAAQAQSGAYTWTVPNPLYFKITNHAQRPFNMNQDIITVQIQFNHNLRSQLDLHKCFLTFKIWTLSHHQNSHFLNLFREHVLMYLDNLGVISINNVIRAVEYVLYDVLEGTEFVQQFTDIKFKLY |
| NCBI Accession | YP_010084311.1 |
|---|---|
| Location | 1211-1618 |
| Gene Name | C2 |
| Protein Name | C2 |
| Coding Region | ATGCAACCTTCGTCACCCTCTACGGCCCACTCTACTCAGGTACCCATCAAAGTCCAGCACAAGATAGGGAAGAAGAGACAACCCCGCAGGAGGAGGATTGATCTAAACTGCGGGTGTTCTATATACGTTAGCTTAGGGTGTGCAAATTATGGATTCACGCACAGGGGTCACCATCACTGCAGCTCAGGCACAGAGTGGCGCGTATACTTGGACAGTGCCAAATCCCCTATATTTCAAAATCACGAATCACGCCCAACGGCCATTCAACATGAATCAGGACATAATCACAGTACAGATACAATTCAACCACAACCTCCGATCTCAGCTGGATTTACACAAGTGCTTCCTGACTTTCAAGATTTGGACTCTCTCACATCATCAGAACTCGCATTTCTTGAATTTATTTAG |
| Protein Sequence | MQPSSPSTAHSTQVPIKVQHKIGKKRQPRRRRIDLNCGCSIYVSLGCANYGFTHRGHHHCSSGTEWRVYLDSAKSPIFQNHESRPTAIQHESGHNHSTDTIQPQPPISAGFTQVLPDFQDLDSLTSSELAFLEFI |
| NCBI Accession | YP_010084312.1 |
|---|---|
| Location | 1518-2609 |
| Gene Name | Rep |
| Protein Name | replication-associated protein |
| Coding Region | ATGCCTACCCCAAATCGTTTTAAAATAAATGCCAAGAATTATTTTCTCACATACCCACACTGCTCTCTTACAAAAGAAGAGGCTCTCTCCCAAATACAAGCCCTACAAACCCCAACTAATAAATTATTCATCCGAATTTGTCGTGAACTACACGAAGATGGGAGCCCTCATCTCCACGTCCTCATCCAATTCGAAGGAAAATATACATGCCGAAATCAACGATTCTTCGATCTCGTATCCCCAACTAGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAGAGCTCGTCAGATGTCAAGACCTACATGGAAAAAGACGGGGACATCCTTGATTTTGGAGTTTTCCAAGTCGATGGAAGATCAGCTAGAGGAGGTTGCCAGTCTGCCAACGACGCATATGCCGAGGCAATCAACTCAGGGTCCAAGTCATCGGCCTTGTCTATATTAAGGGAGAAAGCTCCCAAAGATTATATTTTACAATTTCATAATTTAAATAGTAATTTAGATAGGATTTTTGCTCCTCTGTTGGAGGAATTTGTTTCTCCTTTTTTATCTTCTTCCTTTGATCAAGTTCCAGAGCAACTTGAAGAATGGGCTGCCGAGAACGTCAGGGATTCCGCTGCGCGGCCGTGGAGGCCCATGAGTATTGTGATTGAAGGGGATAGCAGGACAGGGAAGACTATGTGGGCCAGATCTCTACATCCACGTCATAACTACCTTTGCGGCCACCTTGACTTAAGCCCCAAGGTTTACAGCAACGAGGCCTGGTACAACGTCATTGATGACGTGGATCCCCACTACCTAAAACACTTTAAAGAATTCATGGGGGCCCAGAGAGACTGGCAAAGCAACACCAAGTACGGGAAACCAATTCAAATTAAAGGTGGTATCCCAACAATCTTCCTCTGCAATCCAGGCCCAACGTCATCATACACTGAGTATTTAAACGAGGAGAAGAATGCATCTTTGAAACACTGGGCAATTAAAAATGCAACCTTCGTCACCCTCTACGGCCCACTCTACTCAGGTACCCATCAAAGTCCAGCACAAGATAGGGAAGAAGAGACAACCCCGCAGGAGGAGGATTGA |
| Protein Sequence | MPTPNRFKINAKNYFLTYPHCSLTKEEALSQIQALQTPTNKLFIRICRELHEDGSPHLHVLIQFEGKYTCRNQRFFDLVSPTRSAHFHPNIQGAKSSSDVKTYMEKDGDILDFGVFQVDGRSARGGCQSANDAYAEAINSGSKSSALSILREKAPKDYILQFHNLNSNLDRIFAPLLEEFVSPFLSSSFDQVPEQLEEWAAENVRDSAARPWRPMSIVIEGDSRTGKTMWARSLHPRHNYLCGHLDLSPKVYSNEAWYNVIDDVDPHYLKHFKEFMGAQRDWQSNTKYGKPIQIKGGIPTIFLCNPGPTSSYTEYLNEEKNASLKHWAIKNATFVTLYGPLYSGTHQSPAQDREEETTPQEED |
| NCBI Accession | YP_010084313.1 |
|---|---|
| Location | 2159-2581 |
| Gene Name | C4 |
| Protein Name | C4 |
| Coding Region | ATGCCAAGAATTATTTTCTCACATACCCACACTGCTCTCTTACAAAAGAAGAGGCTCTCTCCCAAATACAAGCCCTACAAACCCCAACTAATAAATTATTCATCCGAATTTGTCGTGAACTACACGAAGATGGGAGCCCTCATCTCCACGTCCTCATCCAATTCGAAGGAAAATATACATGCCGAAATCAACGATTCTTCGATCTCGTATCCCCAACTAGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAGAGCTCGTCAGATGTCAAGACCTACATGGAAAAAGACGGGGACATCCTTGATTTTGGAGTTTTCCAAGTCGATGGAAGATCAGCTAGAGGAGGTTGCCAGTCTGCCAACGACGCATATGCCGAGGCAATCAACTCAGGGTCCAAGTCATCGGCCTTGTCTATATTAA |
| Protein Sequence | MPRIIFSHTHTALLQKKRLSPKYKPYKPQLINYSSEFVVNYTKMGALISTSSSNSKENIHAEINDSSISYPQLGQHISIRTFRELRARQMSRPTWKKTGTSLILEFSKSMEDQLEEVASLPTTHMPRQSTQGPSHRPCLY |
References More References in PubMed
| 1 |
Identification of Tomato yellow leaf curl virus Naturally Infecting Common Bean in Japan. Shahid MS, et al. Plant Dis. 2014 Oct;98(10):1447. doi: 10.1094/PDIS-03-14-0316-PDN. PMID: 30703986 |
|---|---|
| 2 |
First Report of Tomato yellow leaf curl virus in China. Wu JB, et al. Plant Dis. 2006 Oct;90(10):1359. doi: 10.1094/PD-90-1359C. PMID: 30780951 |
| 3 |
Complete Genome Sequence of Tomato Leaf Curl New Delhi Virus from Luffa in Indonesia. Wilisiani F, et al. Microbiol Resour Announc. 2019 Apr 11;8(15):e01605-18. doi: 10.1128/MRA.01605-18. PMID: 30975814 |
| 4 |
Kesumawati E, et al. Arch Virol. 2019 Sep;164(9):2379-2383. doi: 10.1007/s00705-019-04316-8. Epub 2019 Jun 15. PMID: 31203434 |
| 5 |
Koeda S, et al. Phytopathology. 2024 Jan;114(1):294-303. doi: 10.1094/PHYTO-04-23-0119-R. Epub 2024 Feb 12. PMID: 37321561 |
| 6 |
Fujiwara A, et al. Microbes Environ. 2025;40(2):ME24095. doi: 10.1264/jsme2.ME24095. PMID: 40307010 |
| 7 |
First Report of Tomato yellow leaf curl virus in Tomato in Costa Rica. Barboza N, et al. Plant Dis. 2014 May;98(5):699. doi: 10.1094/PDIS-08-13-0881-PDN. PMID: 30708517 |
| 8 |
First Report of Tomato yellow leaf curl virus Infecting Eustoma (Eustoma grandiflorum) in Korea. Kil EJ, et al. Plant Dis. 2014 Aug;98(8):1163. doi: 10.1094/PDIS-02-14-0162-PDN. PMID: 30708805 |
| 9 |
Mori T, et al. BMC Res Notes. 2021 Jun 23;14(1):237. doi: 10.1186/s13104-021-05651-3. PMID: 34162412 |
| 10 |
Koeda S, et al. Plant Dis. 2020 Dec;104(12):3221-3229. doi: 10.1094/PDIS-03-20-0613-RE. Epub 2020 Oct 12. PMID: 33044916 |