Tomato leaf curl Gujarat virus
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_000841205.1 |
| Isolate | India: Uttar Pradesh, Varanasi |
| Release date | 2015/2/12 |
| Submitter | Chakraborty,S., Pandey,P.K., Banerjee,M.K., Kalloo,G., Fauquet,C.M. |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
JBrowse
Genome
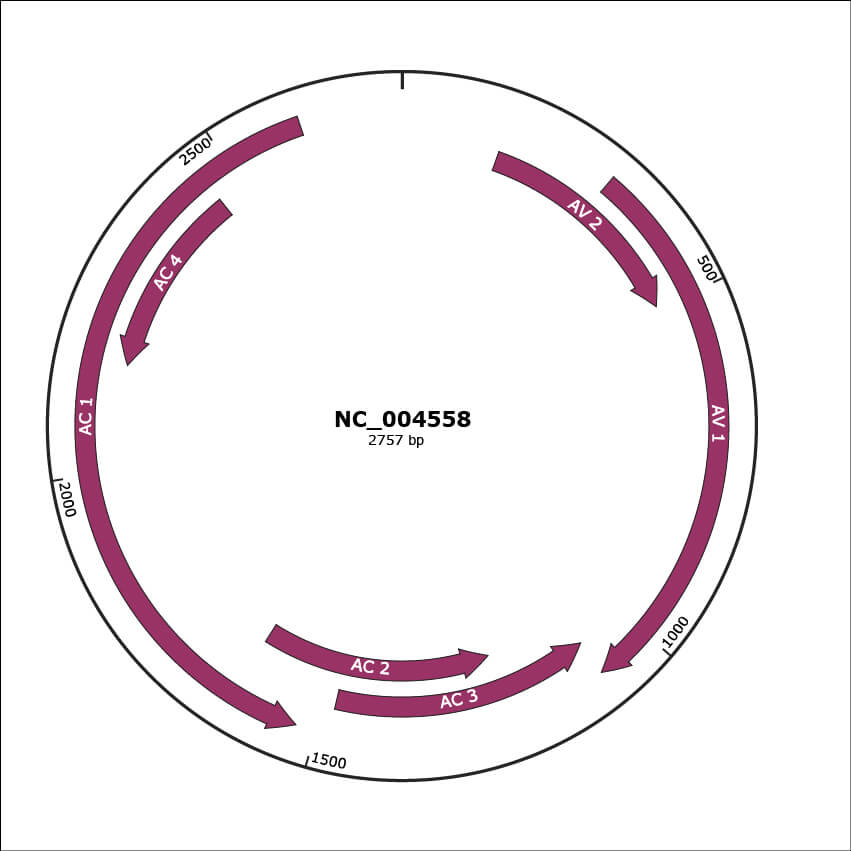
NC_004558
Gene Information
| NCBI Accession | NP_783152.1 |
|---|---|
| Location | 150-497 |
| Gene Name | AV 2 |
| Protein Name | pre-coat protein |
| Coding Region | ATGTGGGATCCTTTGTTAAACGAGTTTCCTGAAACCGTTCACGGTTTTAGATGTATGTTAGCAGTTAAATATCTTCAATTAGTAGAAAAGACTTATTCTCCTGACACAGTCGGTTACGATTTAATCAGGGATTTAATTTCAGTTATTAGGGCTAGAAATTATGTCGAAGCGACCAGCAGATATGCTCATTTTCACGCCCGCTTCGAAGGTACGTCGCCGTCTCAACTTCGACAGCCCATCGGTGAGCCGTGCTGCTGCCCCCATTGTCCGCGTCACCAAAGCAAAGGCATGGGCCAACCGGCCCATGTATCGGAAGCCCAGAATCTACAGAATGTACAGGAGCCCTGA |
| Protein Sequence | MWDPLLNEFPETVHGFRCMLAVKYLQLVEKTYSPDTVGYDLIRDLISVIRARNYVEATSRYAHFHARFEGTSPSQLRQPIGEPCCCPHCPRHQSKGMGQPAHVSEAQNLQNVQEP |
| NCBI Accession | NP_783153.1 |
|---|---|
| Location | 310-1080 |
| Gene Name | AV 1 |
| Protein Name | coat protein |
| Coding Region | ATGTCGAAGCGACCAGCAGATATGCTCATTTTCACGCCCGCTTCGAAGGTACGTCGCCGTCTCAACTTCGACAGCCCATCGGTGAGCCGTGCTGCTGCCCCCATTGTCCGCGTCACCAAAGCAAAGGCATGGGCCAACCGGCCCATGTATCGGAAGCCCAGAATCTACAGAATGTACAGGAGCCCTGATGTGCCTAAGGGCTGTGAGGGTCCGTGTAAGGTCCAATCGTTTGATGCTAAGAACGATATTGGTCACATGGGTAAGGTGATCTGTTTGTCCGATGTGACGAGGGGAATTGGGCTGACCCATCGAGTGGGTAAACGTTTTTGCGTTAAGTCTTTGTATTTCGTCGGCAAGATCTGGATGGACGAGAATATTAAGGTGAAGAACCACACCAACACCGTTATGTTTTGGATAGTACGTGATAGGCGTCCTAGTGGGACTCCCAATGATTTTCAGCAGGTTTTTAACGTATATGATAATGAGCCCTCTACTGCGACTGTTAAGAATGACCAGCGTGATCGCTTTCAAGTCTTACGGAGGTTTCAAGCGACTGTTACTGGAGGACAATATGCAGCCAAGGAGCAGGCGATAATTAGAAGATTTTTTCGTGTTAATAATTACGTAGTTTATAATCACCAGGAAGCTGGGAAGTACGAAAATCATACTGAGAATGCTTTGTTGTTGTATATGGCATGTACTCATGCCTCTAATCCTGTGTATGCGACTCTCAAAGTGAGGAGTTATTTCTATGACTCAGTCACAAACTAA |
| Protein Sequence | MSKRPADMLIFTPASKVRRRLNFDSPSVSRAAAPIVRVTKAKAWANRPMYRKPRIYRMYRSPDVPKGCEGPCKVQSFDAKNDIGHMGKVICLSDVTRGIGLTHRVGKRFCVKSLYFVGKIWMDENIKVKNHTNTVMFWIVRDRRPSGTPNDFQQVFNVYDNEPSTATVKNDQRDRFQVLRRFQATVTGGQYAAKEQAIIRRFFRVNNYVVYNHQEAGKYENHTENALLLYMACTHASNPVYATLKVRSYFYDSVTN |
| NCBI Accession | NP_783154.1 |
|---|---|
| Location | 1077-1481 |
| Gene Name | AC 3 |
| Protein Name | replicational enhancer protein |
| Coding Region | ATGGATTCACGCACAGGGGAATTCATCACTGCAGCTCAAGCAGAGAATGGCGTGTTTATCTGGGAGATTCAAAATCCCCTATATTTCAAGATAACAGAGCACCAAAACCGGCCCTTCCTGATGAACGAAGACATCATCACCGTCCAGATACAGTTCAATCACAACCTGAGGAAAGTGTTGGGGATACACAAATGTTTCCTAACCTACCGATGCTGGATGACCTCACAGCCTCCGACTGGTCGTTTCTTAAGGGTATTTAAAACACAAGTGATGAAATTTCTTAATAATTTGGGTGTAATTAGTTTGAATAATGTAATTAGAGCAGTGAATTATGTACTATGGGATGCATTGACGCAAACAACATTTGTAGACAGTTCAAATATAATAAAATTTAATATTTATTAG |
| Protein Sequence | MDSRTGEFITAAQAENGVFIWEIQNPLYFKITEHQNRPFLMNEDIITVQIQFNHNLRKVLGIHKCFLTYRCWMTSQPPTGRFLRVFKTQVMKFLNNLGVISLNNVIRAVNYVLWDALTQTTFVDSSNIIKFNIY |
| NCBI Accession | NP_783155.1 |
|---|---|
| Location | 1222-1626 |
| Gene Name | AC 2 |
| Protein Name | transcriptional activator protein |
| Coding Region | ATGCAACCTTCGTCACCCTCGAAGGCCCACTCTACTCAAGTACCAATCAAAGTACAGCACAGGCTAGCCAAGAAGGGGACCAGACGTCGACGAGTTGATCTACCGTGTGGCTGTTCTTACTTCATAGCATTAACCTGCAACGACCATGGATTCACGCACAGGGGAATTCATCACTGCAGCTCAAGCAGAGAATGGCGTGTTTATCTGGGAGATTCAAAATCCCCTATATTTCAAGATAACAGAGCACCAAAACCGGCCCTTCCTGATGAACGAAGACATCATCACCGTCCAGATACAGTTCAATCACAACCTGAGGAAAGTGTTGGGGATACACAAATGTTTCCTAACCTACCGATGCTGGATGACCTCACAGCCTCCGACTGGTCGTTTCTTAAGGGTATTTAA |
| Protein Sequence | MQPSSPSKAHSTQVPIKVQHRLAKKGTRRRRVDLPCGCSYFIALTCNDHGFTHRGIHHCSSSREWRVYLGDSKSPIFQDNRAPKPALPDERRHHHRPDTVQSQPEESVGDTQMFPNLPMLDDLTASDWSFLKGI |
| NCBI Accession | NP_783156.1 |
|---|---|
| Location | 1529-2614 |
| Gene Name | AC 1 |
| Protein Name | replicase |
| Coding Region | ATGGCAGCCCCCAATCGCTTTAAAATAAATGCCAAAAATTATTTTCTCACTTATCCCAAGTGCTCTCTAACTAAAGAAGAGGCACTTTCCCAATTATTAAATCTCCAAACACCCACTTTCAAAAAATTTATTAGAATTTGCAGAGAGCTTCACGATGATGGGACTCCTCACTTGCATGTGCTCATCCAGTTCGAAGGAAAGTTCCAATGCAAAAACAATAGATTCTTCGACCTCACATCCCCAACCAGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAGAGCTCAAGCGATGTCAAATCCTACATGGAAAAAGACGGAGACGTCATTGATCATGGAGTTTTCCAAGTCGATGGACGATCAGCTAGAGGAGGTTGCCAATCTGCCAACGACGCATATGCCGAGGCAATCAATTCAGGGTCCAAAGCTCAGGCCCTCAATATATTAAGGGAGAAAGCTCCCAAAGATTTTGTTTTACAATTTCACAATTTAAATTCTAATTTAGATAGGATTTTTACACCTCCAATGGATGTTTATGTTTCACCTTTTATATCTTCTTCTTTCGACCAAGTTCCAGAAGAACTTGAAGCGTGGGCTGCCGAGAACGTCTGCAGTCCCGCTGCGCGGCCTCTGAGACCCATAAGTATAGTAATTGAGGGTGACAGTAGGACGGGGAAGACTATGTGGGCCAGATCATTGGGACCACATAATTATTTGTGTGGTCATCTAGACCTTAGCCCAAAGGTTTACAGTAATGATGCCTGGTACAACGTCATTGATGACGTCGACCCCCACTACCTAAAGCACTTTAAAGAATTCATGGGGGCCCAAAGGGACTGGCAATCAAATACAAAGTACGGGAAGCCAGTTCAAATTAAAGGCGGAATTCCCACTACCTTCCTCTGCAATCCAGGACCCAACTCCAGCTATAAAGAATACTTGGATGAGGAAAAGAATTCTGCACTAAAAAACTGGGCTTTAAAAAATGCAACCTTCGTCACCCTCGAAGGCCCACTCTACTCAAGTACCAATCAAAGTACAGCACAGGCTAGCCAAGAAGGGGACCAGACGTCGACGAGTTGA |
| Protein Sequence | MAAPNRFKINAKNYFLTYPKCSLTKEEALSQLLNLQTPTFKKFIRICRELHDDGTPHLHVLIQFEGKFQCKNNRFFDLTSPTRSAHFHPNIQGAKSSSDVKSYMEKDGDVIDHGVFQVDGRSARGGCQSANDAYAEAINSGSKAQALNILREKAPKDFVLQFHNLNSNLDRIFTPPMDVYVSPFISSSFDQVPEELEAWAAENVCSPAARPLRPISIVIEGDSRTGKTMWARSLGPHNYLCGHLDLSPKVYSNDAWYNVIDDVDPHYLKHFKEFMGAQRDWQSNTKYGKPVQIKGGIPTTFLCNPGPNSSYKEYLDEEKNSALKNWALKNATFVTLEGPLYSSTNQSTAQASQEGDQTSTS |
| NCBI Accession | NP_783157.1 |
|---|---|
| Location | 2164-2460 |
| Gene Name | AC 4 |
| Protein Name | transcriptional regulator protein |
| Coding Region | ATGATGGGACTCCTCACTTGCATGTGCTCATCCAGTTCGAAGGAAAGTTCCAATGCAAAAACAATAGATTCTTCGACCTCACATCCCCAACCAGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAGAGCTCAAGCGATGTCAAATCCTACATGGAAAAAGACGGAGACGTCATTGATCATGGAGTTTTCCAAGTCGATGGACGATCAGCTAGAGGAGGTTGCCAATCTGCCAACGACGCATATGCCGAGGCAATCAATTCAGGGTCCAAAGCTCAGGCCCTCAATATATTAA |
| Protein Sequence | MMGLLTCMCSSSSKESSNAKTIDSSTSHPQPGQHISIRTFRELRAQAMSNPTWKKTETSLIMEFSKSMDDQLEEVANLPTTHMPRQSIQGPKLRPSIY |
References More References in PubMed
| 1 |
Differential pathogenicity among Tomato leaf curl Gujarat virus isolates from India. Ranjan P, et al. Virus Genes. 2013 Dec;47(3):524-31. doi: 10.1007/s11262-013-0977-0. Epub 2013 Sep 12. PMID: 24026875 |
|---|---|
| 2 |
Namgial T, et al. Planta. 2023 Jul 5;258(2):37. doi: 10.1007/s00425-023-04182-4. PMID: 37405593 |
| 3 |
Diverse begomovirus-betasatellite complexes cause tomato leaf curl disease in the western India. Sangeeta, et al. Virus Res. 2023 Apr 15;328:199079. doi: 10.1016/j.virusres.2023.199079. Epub 2023 Mar 3. PMID: 36813240 |
| 4 |
Varun P, et al. 3 Biotech. 2018 May;8(5):243. doi: 10.1007/s13205-018-1254-7. Epub 2018 May 8. PMID: 29744275 |
| 5 |
Prakash V, et al. Planta. 2020 Jul 1;252(1):11. doi: 10.1007/s00425-020-03417-y. PMID: 32613448 |
| 6 |
Chakraborty S, et al. Phytopathology. 2003 Dec;93(12):1485-95. doi: 10.1094/PHYTO.2003.93.12.1485. PMID: 18943612 |
| 7 |
Namgial T, et al. Planta. 2023 Jul 25;258(3):51. doi: 10.1007/s00425-023-04206-z. PMID: 37490148 |
| 8 |
Silencing of tomato CTR1 provides enhanced tolerance against Tomato leaf curl virus infection. Chandan RK, et al. Plant Signal Behav. 2019;14(3):e1565595. doi: 10.1080/15592324.2019.1565595. Epub 2019 Jan 19. PMID: 30661468 |
| 9 |
A New Begomovirus Species Causing Tomato Leaf Curl Disease in Varanasi, India. Chakraborty S, et al. Plant Dis. 2003 Mar;87(3):313. doi: 10.1094/PDIS.2003.87.3.313A. PMID: 30812767 |
| 10 |
First Report of Tomato leaf curl New Delhi virus Infecting Tomato in Bangladesh. Maruthi MN, et al. Plant Dis. 2005 Sep;89(9):1011. doi: 10.1094/PD-89-1011C. PMID: 30786642 |