Tomato enation leaf curl virus
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_000969195.1 |
| Isolate | India: Narsipura, Karnataka |
| Release date | 2015/4/6 |
| Submitter | Swarnalatha,P., Jalali,S., Krishna Reddy,M. |
| Host | |
| Download | Genome |GFF3 |PEP |CDS |
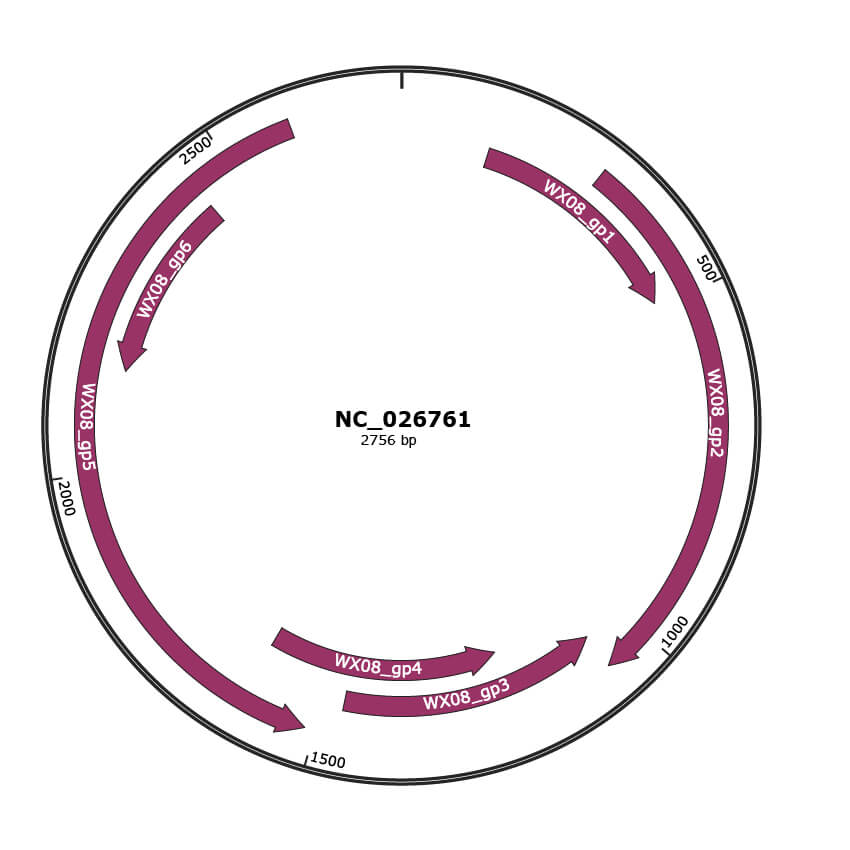
Genomic Organization
JBrowse
Genome
NC_026761
Gene Information
| NCBI Accession | YP_009129274.1 |
|---|---|
| Location | 136-492 |
| Protein Name | AV2 |
| Coding Region | ATGTGGGATCCACTAGTAAACGAGTTTCCTGAAACCGTACACGGTTTTAGATGTATGTTAGCAGTTAAATATCTTCAGTTAGTAGAAAAGACTTATTTTCCTGACACAGTCGGTTACGATTTAATCAGGGATTTAATTTCAGTTATTAGGGCTAGAAATTATGTCGAAGCGACCAGCAGATATAATCATTTCCACTCCCGCTTCGAAGGTACGTCGCCGTCTCAACTTCGACAGCCCATATGTGAGCCGTGCTGTTGCCCCCATTGTCCGCGTCACCAAAGCAAAAGCATGGGCGAACAGGCCCATGAACAGAAAGCCCAGGATGTACAGGATGTACAGAAGTCCAGATGTCCCTAG |
| Protein Sequence | MWDPLVNEFPETVHGFRCMLAVKYLQLVEKTYFPDTVGYDLIRDLISVIRARNYVEATSRYNHFHSRFEGTSPSQLRQPICEPCCCPHCPRHQSKSMGEQAHEQKAQDVQDVQKSRCP |
| NCBI Accession | YP_009129275.1 |
|---|---|
| Location | 296-1066 |
| Protein Name | AV1 |
| Coding Region | ATGTCGAAGCGACCAGCAGATATAATCATTTCCACTCCCGCTTCGAAGGTACGTCGCCGTCTCAACTTCGACAGCCCATATGTGAGCCGTGCTGTTGCCCCCATTGTCCGCGTCACCAAAGCAAAAGCATGGGCGAACAGGCCCATGAACAGAAAGCCCAGGATGTACAGGATGTACAGAAGTCCAGATGTCCCTAGAGGATGTGAAGGCCCATGTAAGGTCCAGTCGTTTGAGTCCAGACACGATGTAGTCCATATAGGCAAGGTCATGTGTATTAGTGATGTCACTCGTGGAACCGGGCTGACCCATAGAGTTGGTAAGCGTTTTTGTGTTAAGTCTGTTTACGTTTTGGGGAAGATATGGATGGATGAGAACATCAAGACCAAGAATCATACGAACAGTGTCATGTTTTTTCTTGTTCGTGACCGTCGTCCTGTTGACAAGCCACAAGACTTTGGAGAGGTGTTCAATATGTTTGACAACGAGCCTAGCACTGCTACTGTTAAGAATATGCACAGAGATCGTTATCAGGTGTTGAGGAAGTGGCATGCAACTGTCACCGGTGGACAGTACGCTTCAAAGGAACAGGCATTAGTGAAGAAGTTTGTTAGGGTTAATAATTATGTTGTTTATAACCAGCAAGAGGCTGGGAAATATGAGAATCATACTGAAAATGCATTGATGTTGTATATGGCGTGTACTCACGCCTCTAACCCTGTGTATGCTACTTTGAAGATACGGATCTATTTTTATGATTCAGTATCAAATTAA |
| Protein Sequence | MSKRPADIIISTPASKVRRRLNFDSPYVSRAVAPIVRVTKAKAWANRPMNRKPRMYRMYRSPDVPRGCEGPCKVQSFESRHDVVHIGKVMCISDVTRGTGLTHRVGKRFCVKSVYVLGKIWMDENIKTKNHTNSVMFFLVRDRRPVDKPQDFGEVFNMFDNEPSTATVKNMHRDRYQVLRKWHATVTGGQYASKEQALVKKFVRVNNYVVYNQQEAGKYENHTENALMLYMACTHASNPVYATLKIRIYFYDSVSN |
| NCBI Accession | YP_009129276.1 |
|---|---|
| Location | 1063-1467 |
| Protein Name | AC3 |
| Coding Region | ATGGATTCACGCACAGCGGAACTCATCAAAGCTTCTCAAGCAGAGAATGGCGTGTTTATCTGGGAGATTCAAAATCCCCTGTATTTCAAGATAACAGAACACAACAACCGGCCATTTCTTATGAAAGAGGACATCATCACCGTCCAAATACAGTTCAATTACAACCTGAGGAAAGCGTTGGAGATACACAAGTGTTTTCTAGCCTACCGAATCTGGATGACTTCACAGCCTCCGACTGGGCAATTCTTAAGGGTCTTTAAGACCCAAGTGCTTAAATATTTAAATAATTTAGGAATTATCAGTATTAATAATGTAATTCGTGCAGTTGATCATGTATTATGGGATGTATTAGAACACATTGTATATGTAGACCAATCTTATTCAATAAAATTCAATATTTATTAA |
| Protein Sequence | MDSRTAELIKASQAENGVFIWEIQNPLYFKITEHNNRPFLMKEDIITVQIQFNYNLRKALEIHKCFLAYRIWMTSQPPTGQFLRVFKTQVLKYLNNLGIISINNVIRAVDHVLWDVLEHIVYVDQSYSIKFNIY |
| NCBI Accession | YP_009129277.1 |
|---|---|
| Location | 1208-1612 |
| Protein Name | AC2 |
| Coding Region | ATGCGATCTTCGTCACCCTCGAAGGCCCACTCTACTCAGGTTCCAATCAAAGTGCAGCACAGGCTAGCCAAGAAGGGGACGAGGCGTCGACGTGTTGATCTCCCTTGTGGGTGTTCATACTTCATAGCATTAGCCTGCCACAACCATGGATTCACGCACAGCGGAACTCATCAAAGCTTCTCAAGCAGAGAATGGCGTGTTTATCTGGGAGATTCAAAATCCCCTGTATTTCAAGATAACAGAACACAACAACCGGCCATTTCTTATGAAAGAGGACATCATCACCGTCCAAATACAGTTCAATTACAACCTGAGGAAAGCGTTGGAGATACACAAGTGTTTTCTAGCCTACCGAATCTGGATGACTTCACAGCCTCCGACTGGGCAATTCTTAAGGGTCTTTAA |
| Protein Sequence | MRSSSPSKAHSTQVPIKVQHRLAKKGTRRRRVDLPCGCSYFIALACHNHGFTHSGTHQSFSSREWRVYLGDSKSPVFQDNRTQQPAISYERGHHHRPNTVQLQPEESVGDTQVFSSLPNLDDFTASDWAILKGL |
| NCBI Accession | YP_009129278.1 |
|---|---|
| Location | 1515-2600 |
| Protein Name | AC1 |
| Coding Region | ATGGTTCCTACAAGACCATTTAGAATAAATGCCAAAAATTATTTTCTCACTTATCCCAAGTGCTCTCTAACAAAAGAGGAAGCACTTTCCCAATTAAAAAACTTAGAAACCCCAACATCCAAAAAATACATCAAGATCTGCAGAGAGCTTCACGAAGATGGGGAACCTCATCTGCACGTGCTCATCCAGTTCGAGGGGAAATACCAGTGCAAGAATCAGAGATTCTTCGACCTGGTATCCCCAACCAGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAATCAAGCTCAGATGTCAAGGCCTACATCGACAAGGACGGGGACACTCTCGAATGGGGAGAGTTTCAGATCGATGGAAGATCAGCAAGAGGAGGACAACAGACAGCAAACGACGCTTACGCCGCAGCACTTAACGCAGGCAGTAAGTCAGAGGCTCTTAGAGTAATAAAGGAACTAGCTCCTAAAGATTTTGTACTACAATTTCATAATTTAAATGCAAATCTAGACAGAATCTTTCAGGAGCCAATGGATGTTTATATTTCTCCTTTTTCTCGTTCTTCTTTTGATCAAGTTCCAGAAGAACTTGAAGTTTGGGCCATCGATAACGTTGTCGATCCCGCTGCGCGGCCTCTGAGGCCTCAAAGTATAGTAATTGAGGGAGACAGTCGTACGGGTAAAACGATGTGGGCTAGGTCATTAGGGCCTCATAATTACCTATGTGGTCATTTAGATCTAAGCCCCAAAGTTTACAGCAATGATGCGTGGTATAACATCATCGATGACGTAGATCCTCACTACCTAAAGCACTTTAAGGAATTCATGGGGGCCCAAAGGGACTGGCAATCAAATACCAAGTACGGAAAGCCAGTTCAAATTAAAGGCGGAATTCCCACTATCTTCCTCTGCAATCCAGGACCTAATTCAAGTTATAAAGAGTTCTTGGATGAAGAGAAGAATAATGCTCTCAAAAATTGGGCTTTAAAGAATGCGATCTTCGTCACCCTCGAAGGCCCACTCTACTCAGGTTCCAATCAAAGTGCAGCACAGGCTAGCCAAGAAGGGGACGAGGCGTCGACGTGTTGA |
| Protein Sequence | MVPTRPFRINAKNYFLTYPKCSLTKEEALSQLKNLETPTSKKYIKICRELHEDGEPHLHVLIQFEGKYQCKNQRFFDLVSPTRSAHFHPNIQGAKSSSDVKAYIDKDGDTLEWGEFQIDGRSARGGQQTANDAYAAALNAGSKSEALRVIKELAPKDFVLQFHNLNANLDRIFQEPMDVYISPFSRSSFDQVPEELEVWAIDNVVDPAARPLRPQSIVIEGDSRTGKTMWARSLGPHNYLCGHLDLSPKVYSNDAWYNIIDDVDPHYLKHFKEFMGAQRDWQSNTKYGKPVQIKGGIPTIFLCNPGPNSSYKEFLDEEKNNALKNWALKNAIFVTLEGPLYSGSNQSAAQASQEGDEASTC |
| NCBI Accession | YP_009129279.1 |
|---|---|
| Location | 2153-2443 |
| Protein Name | AC4 |
| Coding Region | ATGGGGAACCTCATCTGCACGTGCTCATCCAGTTCGAGGGGAAATACCAGTGCAAGAATCAGAGATTCTTCGACCTGGTATCCCCAACCAGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAATCAAGCTCAGATGTCAAGGCCTACATCGACAAGGACGGGGACACTCTCGAATGGGGAGAGTTTCAGATCGATGGAAGATCAGCAAGAGGAGGACAACAGACAGCAAACGACGCTTACGCCGCAGCACTTAACGCAGGCAGTAAGTCAGAGGCTCTTAGAGTAA |
| Protein Sequence | MGNLICTCSSSSRGNTSARIRDSSTWYPQPGQHISIRTFRELNQAQMSRPTSTRTGTLSNGESFRSMEDQQEEDNRQQTTLTPQHLTQAVSQRLLE |
References More References in PubMed
| 1 |
Nagendran K, et al. Virusdisease. 2017 Dec;28(4):425-429. doi: 10.1007/s13337-017-0403-7. Epub 2017 Oct 9. PMID: 29291235 |
|---|---|
| 2 |
Shafiq M, et al. 3 Biotech. 2019 Jun;9(6):204. doi: 10.1007/s13205-019-1727-3. Epub 2019 May 7. PMID: 31139535 |
| 3 |
A distinct seed-transmissible strain of tomato leaf curl New Delhi virus infecting Chayote in India. Sangeetha B, et al. Virus Res. 2018 Oct 15;258:81-91. doi: 10.1016/j.virusres.2018.10.009. Epub 2018 Oct 15. PMID: 30336187 |
| 4 |
Widespread Occurrence of Cotton leaf curl virus on Radish in Pakistan. Mansoor S, et al. Plant Dis. 2000 Jul;84(7):809. doi: 10.1094/PDIS.2000.84.7.809B. PMID: 30832124 |
| 5 |
Ogawa T, et al. Virus Res. 2008 Nov;137(2):235-44. doi: 10.1016/j.virusres.2008.07.021. Epub 2008 Sep 13. PMID: 18722488 |
| 6 |
Genetic Diversity and Pathogenicity Characterization of Tomato-Infecting Begomoviruses in Taiwan. Lai HC, et al. Plant Dis. 2024 Sep;108(9):2688-2700. doi: 10.1094/PDIS-12-22-2937-RE. Epub 2024 Aug 12. PMID: 38587795 |
| 7 |
Venkataravanappa V, et al. Virusdisease. 2022 Jun;33(2):194-207. doi: 10.1007/s13337-022-00772-0. Epub 2022 Jul 1. PMID: 35991698 |
| 8 |
Venkataravanappa V, et al. Microb Pathog. 2023 Jul;180:106127. doi: 10.1016/j.micpath.2023.106127. Epub 2023 Apr 28. PMID: 37119939 |
| 9 |
Iqbal MJ, et al. Virus Res. 2023 Aug;333:199144. doi: 10.1016/j.virusres.2023.199144. Epub 2023 Jun 7. PMID: 37271420 |
| 10 |
Lestari SM, et al. Arch Insect Biochem Physiol. 2023 Feb;112(2):e21984. doi: 10.1002/arch.21984. Epub 2022 Nov 17. PMID: 36397643 |