Tomato chlorotic mottle virus
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_000838345.1 |
| Release date | 2015/2/12 |
| Submitter | Ribeiro,S.G., Lacorte,C.C., Inoue-Nagata,A.K., Carmo,I., Orlandini,D., Andrade,E.C., Nagata,T., Zerbini,F.M. Jr., Zerbini,F.M. |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
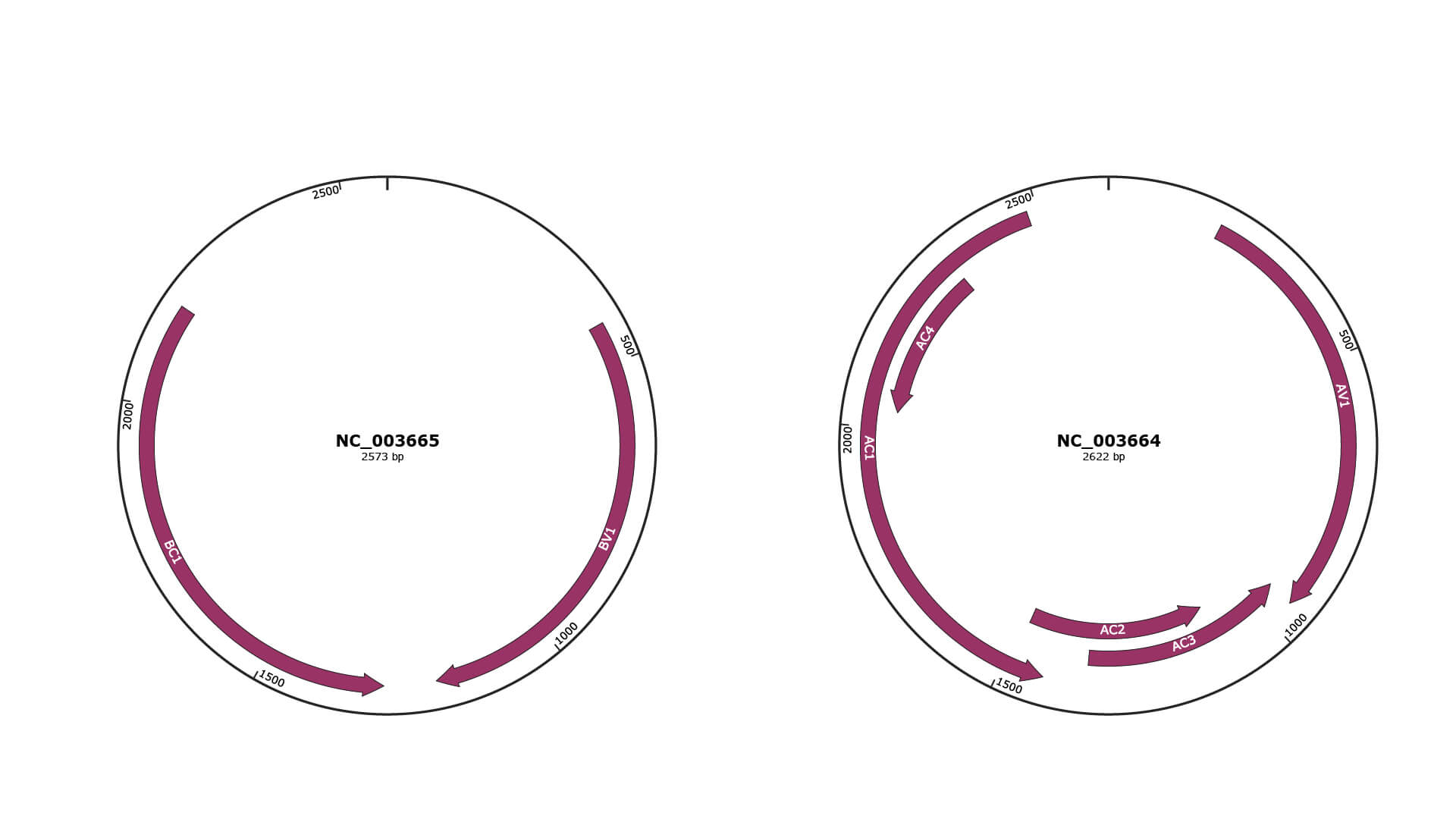
JBrowse
Genome
NC_003665
NC_003664
Gene Information
| NCBI Accession | NP_620014.1 |
|---|---|
| Location | 432-1202 |
| Gene Name | BV1 |
| Protein Name | nuclear shuttle protein |
| Coding Region | ATGTATTTCAATGGTAACAAGCGTCGTTGGTTGTCCACCAATCGCCGTAATTACTTACGATTTCCTACGTTTAAAAGGTCACATGCTGTGGTACGCAATGATGTGAAACGTCGATTTGGTATATCAAACAAGGCACATGACGATAGTAAGATGACCTCTCATAGGATCCACGAAAACCAATATGGGCCTGACTTCGTATTGGGCCATAACTCAGCGCTGTCCACTTTTATAACTTACCCGACTCTTGGTAAAACTGAGCCCAACCGTACTAAATCATACATTAAGTTGAAACGTCTACGTTTTAAAGGTACTGTTAAGATAGAACGTGTTCCTGCTGATTTGAACATGAACGGAATGCCTGCTAAGATAGAGGGCGTATTTTCTCTGGTTATTGTTGTTGATCGTAAACCTCATTTGAGCCCTTCCGGCAGTCTGTACACATTCGATGAGTTATTTGGTGCTAGGATTCATAGCCATGGAAATTTGGCGATAACCCCTTCTCTGATGGATCGTTTTTATATACGCCATGTTGTTAAACGTGTATTGCCTGTGGAGAAAGACAGTAACATGATTGACGTTGATGGGACGACAACATTATCTAGCAGGCGTTATAATTGTTGGGCAAATTTTAGAGACCTCGATCATGAATCATGTAATGGTGTATATGCGAACATAAGCAAGAACGCTCTCTTAGTTTATTATTGTTGGATGTCAGATACCGTTTCCAAAGCATCTACATTTGTATCATTTGACCTTGACTATATTGGATAA |
| Protein Sequence | MYFNGNKRRWLSTNRRNYLRFPTFKRSHAVVRNDVKRRFGISNKAHDDSKMTSHRIHENQYGPDFVLGHNSALSTFITYPTLGKTEPNRTKSYIKLKRLRFKGTVKIERVPADLNMNGMPAKIEGVFSLVIVVDRKPHLSPSGSLYTFDELFGARIHSHGNLAITPSLMDRFYIRHVVKRVLPVEKDSNMIDVDGTTTLSSRRYNCWANFRDLDHESCNGVYANISKNALLVYYCWMSDTVSKASTFVSFDLDYIG |
| NCBI Accession | NP_620015.1 |
|---|---|
| Location | 1293-2174 |
| Gene Name | BC1 |
| Protein Name | movement protein |
| Coding Region | ATGAGTTCTCAGTTGGTTGTTCCTCCAAATGCTTTTAATTATGTTGAGTCGCACAGGGATGAATATCAGCTGTCTCATGACTTGACCGAAATTGTTCTGCAATTTCCTTCTGCGGCGTCGCAGATCACTGCTAAACTTAGTCGCAGCTGTATGAAAATCGACCACTGCGTCATCGAATACAGACAGCAGGTTCCTATAAATGCAACTGGGTCTGTTATAGTTGAAATTCATGACAAACGAATGACCGACAACGAGTCACTGCAGGCGTCATGGACTTTCCCAATTAGATGTAACATAGACTTACATTATTTCTCTTCTTCTTTCTTTTCGCTTAAGGACCCGATTCCATGGAAATTGTACTATAGAGTTAGCGATACGAATGTACACCAAAGGACACATTTCGCCAAATTCAAGGGCAAGTTGAAACTCTCCACAGCAAAACACTCAGTAGACATCCCCTTCCGGGCACCAACAGTTAATATTTTGTCGAAGCAATTCACAGATAAGGATATCGATTTCGTTCATGTAGGTTATGGGTCATGGGAGAGGAAATTACTCAGATGCGCTTCGACAAGAAGCTTTGGGCTTAACGGCCCAATAGAAATTAAAGCAGGTGAGTCATGGGCCTCAAGAAGTACAATTGGTCTCAGCAATTCATGTGTGGACACAGAACGAGGCAACGATCCATATCCATATAAGAACCTAAACAGATTGGATACAGTTATACTAGATCCAGGAGAATCAGCGTCGATCGTTGCGGCACAGAGAACACAGTCCAACATTACTTTATCGTTAGCCCAATTAAATGAAATTGTTAGGACAACAGTACAAGAATGTATTAATACCAATTGTACTCCTTCTGAACCTAAGTCTTTGAAATAA |
| Protein Sequence | MSSQLVVPPNAFNYVESHRDEYQLSHDLTEIVLQFPSAASQITAKLSRSCMKIDHCVIEYRQQVPINATGSVIVEIHDKRMTDNESLQASWTFPIRCNIDLHYFSSSFFSLKDPIPWKLYYRVSDTNVHQRTHFAKFKGKLKLSTAKHSVDIPFRAPTVNILSKQFTDKDIDFVHVGYGSWERKLLRCASTRSFGLNGPIEIKAGESWASRSTIGLSNSCVDTERGNDPYPYKNLNRLDTVILDPGESASIVAAQRTQSNITLSLAQLNEIVRTTVQECINTNCTPSEPKSLK |
| NCBI Accession | NP_620009.1 |
|---|---|
| Location | 199-954 |
| Gene Name | AV1 |
| Protein Name | capsid protein |
| Coding Region | ATGCCTAAGCGCGATGCCCCATGGCGCCACATGGCAAGTACGTCTAAGATTAGCCGGGGTGTTAATAATTCTCCTCGGGCAGGAGTTGGGCCAAAATCCAATAAGGCCGCTGATTGGGTAAACAGGCCCATGTACAGGAAGCCCAAGATATACAGGATGTACAGAACCCCCGATGTTCCAAGGGGCTGTGAAGGCCCATGTAAAGTCCAATCGTTTGAACAGCGTCACGACATCTCCCATACTGGGAAGGTGATGTGCGTCTCTGATGTGACACGTGGTAACGGTATTACACACCGTGTGGGTAAGCGTTTCTGTGTTAAGTCTGTGTATATACTTGGTAAGATATGGATGGATGAGAACATCAAGTTGAAGAACCACACAAACAGTGCTATGTTTTGGTTGGTCAGGGATCGTAGACCGTATGGTACCCCAATGGATTTTGGTCAGGTGTTCAACATGTTCGACAACGAGCCCAGTACTGCCACCGTGAAGAACGATCTTCGTGATCGTTTTCAAGTTATGCACAAGTTTTACGCCAAGGTTACGGGTGGACAATACGCAAGTAACGAACAGGCGCTGGTCAAGCGGTTCTGGAAGGTCAACAATCACGTCGTTTACAACCATCAAGAAGCCGGGAAGTACGAGAATCATACTGAGAACGCACTATTACTGTATATGGCATGTACTCATGCCTCTAACCCAGTGTATGCAACTTTAAAAATTCGGATCTATTTTTATGATTCGATAACAAATTAA |
| Protein Sequence | MPKRDAPWRHMASTSKISRGVNNSPRAGVGPKSNKAADWVNRPMYRKPKIYRMYRTPDVPRGCEGPCKVQSFEQRHDISHTGKVMCVSDVTRGNGITHRVGKRFCVKSVYILGKIWMDENIKLKNHTNSAMFWLVRDRRPYGTPMDFGQVFNMFDNEPSTATVKNDLRDRFQVMHKFYAKVTGGQYASNEQALVKRFWKVNNHVVYNHQEAGKYENHTENALLLYMACTHASNPVYATLKIRIYFYDSITN |
| NCBI Accession | NP_620010.1 |
|---|---|
| Location | 951-1349 |
| Gene Name | AC3 |
| Protein Name | replication-enhancer protein |
| Coding Region | ATGGATTCACGCACAGGGGAACTCATCACTGCACATCAGGCAGAGAATGGCGTGTATATCTGGGAAATATCAAATCCCCTTTATTTCAAGATGTACAATGTCGAGGACATACTGTACACGAGGACCAGAGTGTACCACGTACAGATCCGGTTCAATCACAACCTGAGGAGAGTACTGGGTCTTCACAAGGCTTATCTCAACTTCCAAATCTGGACGACATCCCTGAGAGCTTCTGGGATGACATATTTAAATAGATTTAAATATTTAGTAGTGTTGTATATTGATCAGTTAGGCGTTATTTCAATTAATAATGTAATTAGAGCTGTTCGATTCGCAACAGACAGACAATATGTAAATGCTGTACTCGAAGATCATTCAATAAAATACAAACTTTATTAA |
| Protein Sequence | MDSRTGELITAHQAENGVYIWEISNPLYFKMYNVEDILYTRTRVYHVQIRFNHNLRRVLGLHKAYLNFQIWTTSLRASGMTYLNRFKYLVVLYIDQLGVISINNVIRAVRFATDRQYVNAVLEDHSIKYKLY |
| NCBI Accession | NP_620011.1 |
|---|---|
| Location | 1096-1485 |
| Gene Name | AC2 |
| Protein Name | trans-activation protein |
| Coding Region | ATGCGCAATTCATCTTTCTCAACTCCCCCCTCTATCAAAGTTCAACATCGGGCTGCTAAAAAGAGAGCTATCCGAAGAAGACGAATTGATTTGCAGTGCGGGTGCTCCATTTTCGTACACATTGACTGCGCAGGACATGGATTCACGCACAGGGGAACTCATCACTGCACATCAGGCAGAGAATGGCGTGTATATCTGGGAAATATCAAATCCCCTTTATTTCAAGATGTACAATGTCGAGGACATACTGTACACGAGGACCAGAGTGTACCACGTACAGATCCGGTTCAATCACAACCTGAGGAGAGTACTGGGTCTTCACAAGGCTTATCTCAACTTCCAAATCTGGACGACATCCCTGAGAGCTTCTGGGATGACATATTTAAATAG |
| Protein Sequence | MRNSSFSTPPSIKVQHRAAKKRAIRRRRIDLQCGCSIFVHIDCAGHGFTHRGTHHCTSGREWRVYLGNIKSPLFQDVQCRGHTVHEDQSVPRTDPVQSQPEESTGSSQGLSQLPNLDDIPESFWDDIFK |
| NCBI Accession | NP_620012.1 |
|---|---|
| Location | 1427-2482 |
| Gene Name | AC1 |
| Protein Name | replication-associated protein |
| Coding Region | ATGCCACTCCCCAGACATTTCCGAGTAAATTCCAAAAATTATTTTCTAACATATCCTCACTGCTCTCTTTCCAAAGAAGAAGCACTTTCCCAATTACTAGCTCTGAACACACCAACAAACAAATTATTCATTCGTGTATCAAGAGAACTACACGAAAATGGGGAACCTCATCTCCACGTGCTCATCCAGTTCGAAGGTAAATACAACTGCAAGAACAACAAGTTCTTCGACCTCGTATCCCCAACCAGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAGAGCTCCTCAGACGTCAAATCATACGTGGAGAAAGACGGAGACTTCGTTGATCATGGAGTTTTCCAGATCGATGGAAGATCAGCAAGAGGAGGTCAGCAATCTGCCAACGACACGTACGCCAAGGTTCTCAACGCAGGATCAGTCATGGAAGCACTCAATATATTAAGAGAGGAACAACCCAAAGATTTCGTGCTTCAACATCATAATATTAAATCTAATCTAGAGCGTATTTTCGTTAAGGCTCCAGAACCTTGGGTTCCTCCGTTTCCCCTCTCATCGTTCACTAACGTTCCTGTCGAGATGCAAGACTGGGTCAATGATTATTTCGGGAGAGATGCCGCTGCGCGGCCGGAGAGACCTATCAGTATAATAATCGAGGGTGATTCAAGGACGGGGAAGGCAATGTGGGCACGTACATTGGGGTCCCATAATTACTTGAGTGGTCACCTCGATTTCAATTCAAAGGTCTACTCAAACGATGTGCAGTATAACGTCATTGATGATGTCGCACCGCATTATCTAAAACTAAAGCACTGGAAAGAATTGATTGGGGCCCAAAGGGATTGGCAATCAAACTGCAAGTACGGAAAGCCGGTTCAAATTAAAGGTGGTATCCCATGCATCGTGCTTTGCAATCCTGGCGAGGGGGCCAGCTATAAATGTTTCCTAAACAAACAGGAAAACTCAGCATTAGATAGTTGGACAAAGCACAATGCGCAATTCATCTTTCTCAACTCCCCCCTCTATCAAAGTTCAACATCGGGCTGCTAA |
| Protein Sequence | MPLPRHFRVNSKNYFLTYPHCSLSKEEALSQLLALNTPTNKLFIRVSRELHENGEPHLHVLIQFEGKYNCKNNKFFDLVSPTRSAHFHPNIQGAKSSSDVKSYVEKDGDFVDHGVFQIDGRSARGGQQSANDTYAKVLNAGSVMEALNILREEQPKDFVLQHHNIKSNLERIFVKAPEPWVPPFPLSSFTNVPVEMQDWVNDYFGRDAAARPERPISIIIEGDSRTGKAMWARTLGSHNYLSGHLDFNSKVYSNDVQYNVIDDVAPHYLKLKHWKELIGAQRDWQSNCKYGKPVQIKGGIPCIVLCNPGEGASYKCFLNKQENSALDSWTKHNAQFIFLNSPLYQSSTSGC |
| NCBI Accession | NP_620013.1 |
|---|---|
| Location | 2032-2325 |
| Gene Name | AC4 |
| Protein Name | putative AC4 protein |
| Coding Region | ATGGGGAACCTCATCTCCACGTGCTCATCCAGTTCGAAGGTAAATACAACTGCAAGAACAACAAGTTCTTCGACCTCGTATCCCCAACCAGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAGAGCTCCTCAGACGTCAAATCATACGTGGAGAAAGACGGAGACTTCGTTGATCATGGAGTTTTCCAGATCGATGGAAGATCAGCAAGAGGAGGTCAGCAATCTGCCAACGACACGTACGCCAAGGTTCTCAACGCAGGATCAGTCATGGAAGCACTCAATATATTAA |
| Protein Sequence | MGNLISTCSSSSKVNTTARTTSSSTSYPQPGQHISIRTFRELRAPQTSNHTWRKTETSLIMEFSRSMEDQQEEVSNLPTTRTPRFSTQDQSWKHSIY |
References More References in PubMed
| 1 |
Carmo LS, et al. J Proteomics. 2017 Jan 16;151:284-292. doi: 10.1016/j.jprot.2016.07.018. Epub 2016 Jul 22. PMID: 27457268 |
|---|---|
| 2 |
Ribeiro SG, et al. J Virol. 2007 Feb;81(4):1563-73. doi: 10.1128/JVI.01238-06. Epub 2006 Nov 29. PMID: 17135316 |
| 3 |
Ribeiro SG, et al. Phytopathology. 2007 Jun;97(6):702-11. doi: 10.1094/PHYTO-97-6-0702. PMID: 18943601 |
| 4 |
Fontenelle MR, et al. Virus Res. 2007 Jun;126(1-2):262-7. doi: 10.1016/j.virusres.2007.02.009. Epub 2007 Mar 26. PMID: 17367887 |
| 5 |
Carmo LS, et al. Proteomics. 2013 Jun;13(12-13):1947-60. doi: 10.1002/pmic.201200547. Epub 2013 May 3. PMID: 23533094 |
| 6 |
Lacerda AL, et al. PLoS One. 2015 Aug 28;10(8):e0136820. doi: 10.1371/journal.pone.0136820. eCollection 2015. PMID: 26317870 |
| 7 |
Potatovirus X and Tobacco mosaic virus-based vectors compatible with the Gateway cloning system. Lacorte C, et al. J Virol Methods. 2010 Mar;164(1-2):7-13. doi: 10.1016/j.jviromet.2009.11.005. Epub 2009 Nov 10. PMID: 19903495 |
| 8 |
Galvão RM, et al. J Gen Virol. 2003 Mar;84(Pt 3):715-726. doi: 10.1099/vir.0.18783-0. PMID: 12604824 |