Tobacco mottle leaf curl virus
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_002986925.1 |
| Isolate | Cuba:Sancti Spiritus |
| Release date | 2018/8/26 |
| Submitter | Dominguez,M., Ramos,P.L., Sanchez,Y., Crespo,J., Andino,V., Pujol,M., Borroto,C., Milagros,D.M. |
| Download | Genome |GFF3 |PEP |CDS |
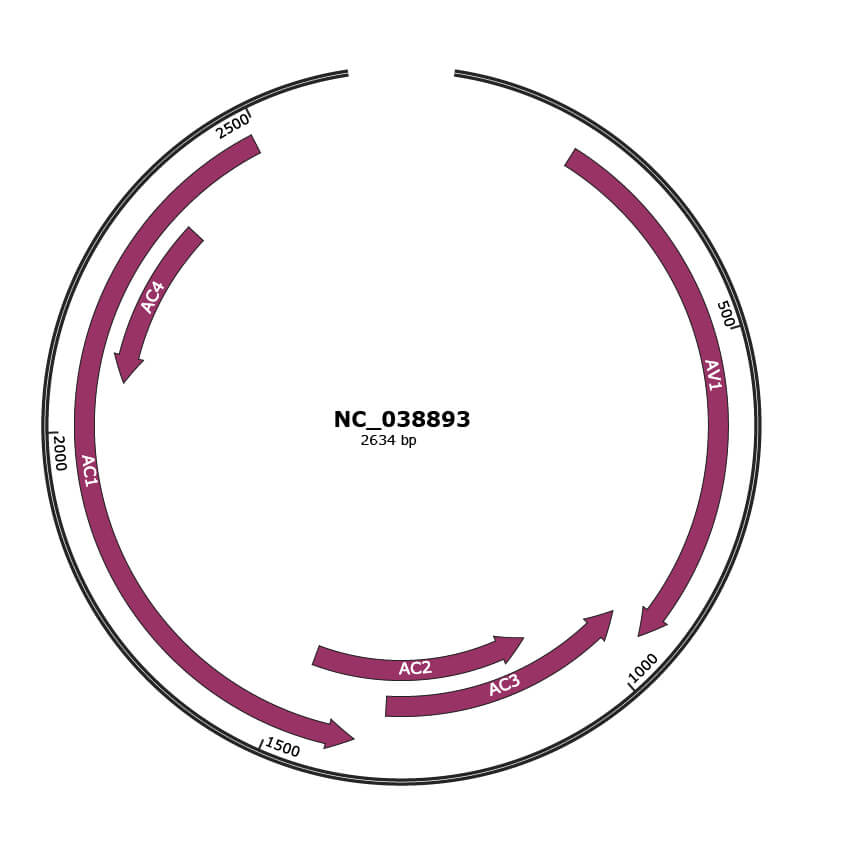
Genomic Organization
JBrowse
Genome
NC_038893
Gene Information
| NCBI Accession | YP_009508169.1 |
|---|---|
| Location | 182-946 |
| Gene Name | AV1 |
| Protein Name | coat protein |
| Coding Region | ATGCCTAAGCGCGATCTCCCATGGCGCCCTTACGCGAAAATATCCAAGGTGAGCCGCAATGCGAATTACTCGCCCCGTACAAGAGTTGGCCCAATTGTCGACAAGGCCGCTGAATGGGTGAACAGGCCCATGTATAGGAAGCCCAGGATCTATCGGACGGTGAGAACCCCCGATGTGCCCAGGGGATGTGAAGGGCCTTGTAAGGTCCAGTCCTATGAACAGCGCCACGACATCTCGCACGTGGGCAAGGTCATGTGTATATCCGACGTGACACGTGGTAATGGCATTACCCATCGTGTTGGTAAGCGTTTTTGTGTTAAGTCTGTGTATATCCTAGGGAAGATCGGGAAAATTTGGATGGATGAGAACATCAAGCTCAAGAACCACACGAACAGCGTCATGTTCTGGTTGGTCAGGGACCGTAGACCGTATGGAACGCCGATGGATTTCGGTCAGGTGTTCAACATGTTCGACAACGAGCCCAGTACCGCTACGGTTAAGAACGATCTACGTGATCGTTACCAGGTCATGCATCGGTTCCATGCTAAGGTCACGGGTGGACAGTATGCCAGCAACGAGCAGGCGATTGTGAGGCGGTTTTGGAAGGTCAACAACCACGTGGTCTACAACCACCAGGAGGCTGGCAAGTACGAGAACCACACGGAGAACGCCCTGTTATTGTATATGGCATGTACTCATGCCTCTAATCCTGTATATGCAACTCTGAAGATACGAATCTATTTTTATGATTCGGTCATGAATTAA |
| Protein Sequence | MPKRDLPWRPYAKISKVSRNANYSPRTRVGPIVDKAAEWVNRPMYRKPRIYRTVRTPDVPRGCEGPCKVQSYEQRHDISHVGKVMCISDVTRGNGITHRVGKRFCVKSVYILGKIGKIWMDENIKLKNHTNSVMFWLVRDRRPYGTPMDFGQVFNMFDNEPSTATVKNDLRDRYQVMHRFHAKVTGGQYASNEQAIVRRFWKVNNHVVYNHQEAGKYENHTENALLLYMACTHASNPVYATLKIRIYFYDSVMN |
| NCBI Accession | YP_009508170.1 |
|---|---|
| Location | 943-1341 |
| Gene Name | AC3 |
| Protein Name | replication enhancement protein |
| Coding Region | ATGGATTCACGCACCGGGGAACCCATCACTGCACATCAGGCGGAGAATGGCGCGTATATCTGGGAGATAGAAAATCCCCTGTATTTCAAGATATACAACGTAGAGGACCTTCCATACACCAGGACACGAATATACCACATCCAGATACGGTTCAACCACAACCTCAGGAGAGCGTTGGATCTCCACAAGGCTTACCTGAACTTCCAAGTCTGGACGACATCGACGACAGCTTCTGGAACGACATATTTGCTTAGGTTTAGGCACTTAGTTAACATGTACTTAGATCAATTAGGCGTGATTTCCATTAACAATGTAATTAGAGCTGTTCGTTTCGCGACAGACAGACCATATGTAAATCATGTACTGGAAAATCACTCAATAAAATTCAAAATTTATTAA |
| Protein Sequence | MDSRTGEPITAHQAENGAYIWEIENPLYFKIYNVEDLPYTRTRIYHIQIRFNHNLRRALDLHKAYLNFQVWTTSTTASGTTYLLRFRHLVNMYLDQLGVISINNVIRAVRFATDRPYVNHVLENHSIKFKIY |
| NCBI Accession | YP_009508171.1 |
|---|---|
| Location | 1088-1474 |
| Gene Name | AC2 |
| Protein Name | transactivator protein |
| Coding Region | ATGCGGCTCGTCCCCTCCAGCCCCCCTCTATCAAGAAGGCACACAGGCAAGCCAGAGAGGAGGGCCATCAGGAGGAGACGCATTGATCTAGAGTGCGGGTGCTCCATATACTTCCACATAGACTGCCGGGGACATGGATTCACGCACCGGGGAACCCATCACTGCACATCAGGCGGAGAATGGCGCGTATATCTGGGAGATAGAAAATCCCCTGTATTTCAAGATATACAACGTAGAGGACCTTCCATACACCAGGACACGAATATACCACATCCAGATACGGTTCAACCACAACCTCAGGAGAGCGTTGGATCTCCACAAGGCTTACCTGAACTTCCAAGTCTGGACGACATCGACGACAGCTTCTGGAACGACATATTTGCTTAG |
| Protein Sequence | MRLVPSSPPLSRRHTGKPERRAIRRRRIDLECGCSIYFHIDCRGHGFTHRGTHHCTSGGEWRVYLGDRKSPVFQDIQRRGPSIHQDTNIPHPDTVQPQPQESVGSPQGLPELPSLDDIDDSFWNDIFA |
| NCBI Accession | YP_009508172.1 |
|---|---|
| Location | 1384-2490 |
| Gene Name | AC1 |
| Protein Name | replication associated protein |
| Coding Region | ATGCCATCGGTTAAGAAATTTAGAGTTCAGTCAAGAAATTATTTTCTCACTTATCCACAGTGCTCTCTTAGCAAAGAGGAGGCACTTTCCCAATTACAAAACCTAGACACACCAGTAAACAAGAAGTTCATCAAGATATGCAGAGAGCTTCATGAGAATGGGGAGCCTCATCTCCACGTGCTTGTCCAATTCGAAGGCAAGTACGTCTGCACGAATTACAGATTCTTCGACTTGGTCTCCCCATCTCGGTCAGCACATTTCCATCCGAACGTTCAGGGAGCTAAATCGAGCTCCGACGTCAAGTCCTACATCGACAAGGACGGAGATACACTGGAATGGGGAGAGTTCCAGGTCGACGGAAGAAGTGCTAGAGGAGGCTGCCAATCGGCTAACGACACGTATGCCAAGGCGTTAAATGCCTCTTCTGCAGAAGAGGCTCTACAAATAATAAAAGAAGAGCAGCCTCAACATTTCTTCCTCCAACATCATAACCTCGTCGCAAATGCACAAAGGATCTTTCAAAAGGCTCCGGAACCGTGGGTCCCTCCGTTTCAACTCTCCTCGCTCACTAACGTCCCGGAAGAAATGCAAGAGTGGGCCGATGAGTATTTTGGAAGGGGTTCCGCTGCGCGGCCGGAGAGACCTATGAGTATCATCGTCGAAGGTGATTCAAGGACCGGGAAGACAATGTGGGCCTGCAGTAAGACAATGTGGGCACGTGCGTTAGGCCCACATAACTATCTCTCTGGTCACCTAGACTTCAATCACCGGGTCTACTCGAACGAAGTGGAGTATAACGTAATTGATGACGTCGCACCGCACTATCTAAAGCTAAAGCACTGGAAAGAATTGCTGGGGTCACAAAAAGACTGGCAGTCAAACTGCAAATACGGAAAGCCAGTTCAAATTAAAGGCGGGATCCCAGCAATCGTGCTATGCAATCCTGGTGAGGGTGCCAGCTATAAAGATTTCCTAGACAAAGAGGAAAACTCATCACTCAGGAATTTTGGGGACAGATGCGGCTCGTCCCCTCCAGCCCCCCTCTATCAAGAAGGCACACAGGCAAGCCAGAGAGGAGGGCCATCAGGAGGAGACGCATTGATCTAG |
| Protein Sequence | MPSVKKFRVQSRNYFLTYPQCSLSKEEALSQLQNLDTPVNKKFIKICRELHENGEPHLHVLVQFEGKYVCTNYRFFDLVSPSRSAHFHPNVQGAKSSSDVKSYIDKDGDTLEWGEFQVDGRSARGGCQSANDTYAKALNASSAEEALQIIKEEQPQHFFLQHHNLVANAQRIFQKAPEPWVPPFQLSSLTNVPEEMQEWADEYFGRGSAARPERPMSIIVEGDSRTGKTMWACSKTMWARALGPHNYLSGHLDFNHRVYSNEVEYNVIDDVAPHYLKLKHWKELLGSQKDWQSNCKYGKPVQIKGGIPAIVLCNPGEGASYKDFLDKEENSSLRNFGDRCGSSPPAPLYQEGTQASQRGGPSGGDALI |
| NCBI Accession | YP_009508173.1 |
|---|---|
| Location | 2076-2339 |
| Gene Name | AC4 |
| Protein Name | AC4 protein |
| Coding Region | ATGAGAATGGGGAGCCTCATCTCCACGTGCTTGTCCAATTCGAAGGCAAGTACGTCTGCACGAATTACAGATTCTTCGACTTGGTCTCCCCATCTCGGTCAGCACATTTCCATCCGAACGTTCAGGGAGCTAAATCGAGCTCCGACGTCAAGTCCTACATCGACAAGGACGGAGATACACTGGAATGGGGAGAGTTCCAGGTCGACGGAAGAAGTGCTAGAGGAGGCTGCCAATCGGCTAACGACACGTATGCCAAGGCGTTAA |
| Protein Sequence | MRMGSLISTCLSNSKASTSARITDSSTWSPHLGQHISIRTFRELNRAPTSSPTSTRTEIHWNGESSRSTEEVLEEAANRLTTRMPRR |
References More References in PubMed
| 1 |
Prasanth G, et al. Plant Dis. 2008 Feb;92(2):311. doi: 10.1094/PDIS-92-2-0311B. PMID: 30769415 |
|---|---|
| 2 |
Zhou YC, et al. Arch Virol. 2008;153(4):693-706. doi: 10.1007/s00705-008-0042-9. Epub 2008 Feb 16. PMID: 18278427 |
| 3 |
Pest categorisation of non-EU viruses of Rubus L. EFSA Panel on Plant Health (PLH), et al. EFSA J. 2020 Jan 16;18(1):e05928. doi: 10.2903/j.efsa.2020.5928. eCollection 2020 Jan. PMID: 32626483 |
| 4 |
Jyothsna P, et al. Arch Virol. 2013 Jan;158(1):217-24. doi: 10.1007/s00705-012-1468-7. Epub 2012 Sep 15. PMID: 22983111 |
| 5 |
Zhang X, et al. PLoS One. 2023 Jan 25;18(1):e0280303. doi: 10.1371/journal.pone.0280303. eCollection 2023. PMID: 36696381 |
| 6 |
Sardo L, et al. Virus Res. 2011 Nov;161(2):170-80. doi: 10.1016/j.virusres.2011.07.021. Epub 2011 Aug 4. PMID: 21843560 |
| 7 |
Molecular characterization of a new begomovirus infecting Mirabilis jalapa in northern India. Kulshreshtha A, et al. Arch Virol. 2017 Jul;162(7):2163-2167. doi: 10.1007/s00705-017-3330-4. Epub 2017 Mar 24. PMID: 28342034 |
| 8 |
Ringeera KH, et al. Mol Biol Rep. 2025 Sep 13;52(1):901. doi: 10.1007/s11033-025-11020-1. PMID: 40944776 |
| 9 |
Kumari N, et al. Virusdisease. 2020 Sep;31(3):323-332. doi: 10.1007/s13337-020-00597-9. Epub 2020 May 15. PMID: 32904916 |
| 10 |
Rapid LAMP-Based Detection of Mixed Begomovirus Infections in Field-Grown Tomato Plants. Ruiz-Otaño Y, et al. Viruses. 2025 Dec 23;18(1):19. doi: 10.3390/v18010019. PMID: 41600784 |