Rose leaf curl virus
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_000921455.1 |
| Isolate | India |
| Release date | 2015/2/22 |
| Submitter | Nehra,C., Sahu,A.K., Gaur,R.K. |
| Host | |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
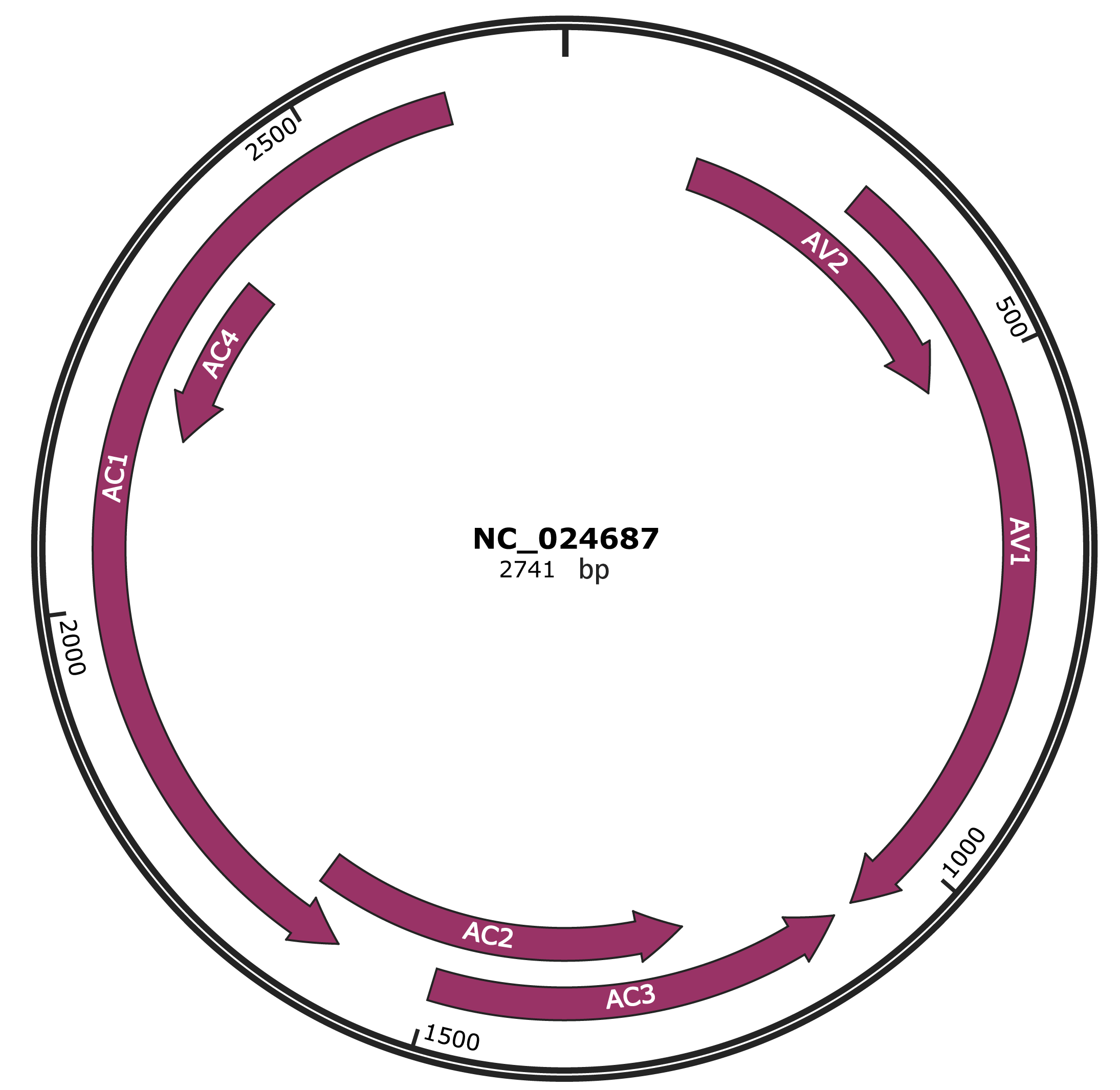
JBrowse
Genome
NC_024687
Gene Information
| NCBI Accession | YP_009051687.1 |
|---|---|
| Location | 144-509 |
| Gene Name | AV2 |
| Protein Name | pre-coat protein |
| Coding Region | ATGTGGGACCCACTATTGAATGATTTTCCAGAAACTGTTCATGGGTTTAGGTGTATGCTAGCAATTAAATACTTGCAGCTAGTAAAAAATACGTTTTCCCCAGACACTCTGGGATACGATTTAATTGGGGATTTGATTTCAGTAATAAGGGCTAGGGATTATGTCGAAGCGACCCGCAGATATAATCATTTCCACGCCCGCCTCGAAGGTACGCCCCCGTCTCAACTTCGACAGCCCATATGCGAGCCGTGCTGCTCCCCCCATTGTCCGCGTCACAAAGGCAAGGGCATGGGCGAACAGGCCCATGAACAGAAAGCCCAGGATGTACAGGATGTACAGAAGCCCAGATGTTCCGAGGGGATGTAA |
| Protein Sequence | MWDPLLNDFPETVHGFRCMLAIKYLQLVKNTFSPDTLGYDLIGDLISVIRARDYVEATRRYNHFHARLEGTPPSQLRQPICEPCCSPHCPRHKGKGMGEQAHEQKAQDVQDVQKPRCSEGM |
| NCBI Accession | YP_009051688.1 |
|---|---|
| Location | 304-1074 |
| Gene Name | AV1 |
| Protein Name | coat protein |
| Coding Region | ATGTCGAAGCGACCCGCAGATATAATCATTTCCACGCCCGCCTCGAAGGTACGCCCCCGTCTCAACTTCGACAGCCCATATGCGAGCCGTGCTGCTCCCCCCATTGTCCGCGTCACAAAGGCAAGGGCATGGGCGAACAGGCCCATGAACAGAAAGCCCAGGATGTACAGGATGTACAGAAGCCCAGATGTTCCGAGGGGATGTAAAGGCCCATGCAAGGTCCAGTCATTTGAGTCCAGACATGATATCCAGCACATTGGTAAAGTCATGTGTGTTAGTGATGTTACTCGTGGTATTGGGCTGACCCACAGGGTTGGCAAGAGGTTCTGTGTTAAGTCCGTTTATGTTCTGGGCAAGATCTGGATGGATGAGAACAACAAGACTAAGAATCATACGAATAGTGTTATGTTTTTCCTTGTTAGGGATCGTAGGCCTGTTGACAAGCCTCAAGTTTTTGGTGAGGTGTTTAACATGTTTGATAATGAGCCCAGCACGGGGACTGTGAAGAATGTTCATCGTGATAGGTACCAGGTATTAAGGAAGTGGGACGCAACTGTGACAGGTGGCCTGTATGCATCGAAGGAGCAGGCTCTCGTGAAGAAGTTTATTAGGGTTAATAATTATTTTGTGTACAACCAGCAAGAGGCTGGCAAGTATGAGAATCATACTGAGAATGCATTGATGTTGTATATGGCGTGTACCCACGCCTCTAACCCTGTGTATGCCACACTGAAGATACGGATCTATTTTTATGATTCAGTATCGAATTAA |
| Protein Sequence | MSKRPADIIISTPASKVRPRLNFDSPYASRAAPPIVRVTKARAWANRPMNRKPRMYRMYRSPDVPRGCKGPCKVQSFESRHDIQHIGKVMCVSDVTRGIGLTHRVGKRFCVKSVYVLGKIWMDENNKTKNHTNSVMFFLVRDRRPVDKPQVFGEVFNMFDNEPSTGTVKNVHRDRYQVLRKWDATVTGGLYASKEQALVKKFIRVNNYFVYNQQEAGKYENHTENALMLYMACTHASNPVYATLKIRIYFYDSVSN |
| NCBI Accession | YP_009051689.1 |
|---|---|
| Location | 1095-1499 |
| Gene Name | AC3 |
| Protein Name | replication enhancer protein |
| Coding Region | ATGGATTCACGCACAGGGGAATCCATCACTGCAGCTCAAGCACAGAATGGGGTGTTTATCTGGGAGGTTCAAAATCCCCTCTATTTCAAGATAACAGAGCACCACAACAGGGGATTCCTCATGAACCAAGACATCATCACTGCCCAGATACAGTTCAATCACAACCTGAGGAAAGCGTTGGGAATACACAAATGTTTCCTAGCCTTCCGAATCTGGATGACCTCACAGCCTCCGACTGGTCGTTTCTTAAGGGTCTTTAAGACACAAGTCCTTAAGTATTTAGATAATCTAGGAGTTATCAGTATTAACAAAGTAATTAGAGCAGTTGATCATGTATTATGGAATGTATTAGAGCATATTGTATATGTAAAACAATCATCTTCAATAAAATTCAATATTTATTAA |
| Protein Sequence | MDSRTGESITAAQAQNGVFIWEVQNPLYFKITEHHNRGFLMNQDIITAQIQFNHNLRKALGIHKCFLAFRIWMTSQPPTGRFLRVFKTQVLKYLDNLGVISINKVIRAVDHVLWNVLEHIVYVKQSSSIKFNIY |
| NCBI Accession | YP_009051690.1 |
|---|---|
| Location | 1240-1647 |
| Gene Name | AC2 |
| Protein Name | transcriptional activator protein |
| Coding Region | ATGCAACATTCATCACCCTCACAGAGCCACTGTACTCAGGTGCCCATTAAGGTCCAACACAGGCTGGCCAAGCGTAGAGCCATCCGGCGTAAGAGGGTTGACCTCGAGTGTGGCTGCCCAAACTACGTACACATCAACCCCCACAACCATGGATTCACGCACAGGGGAATCCATCACTGCAGCTCAAGCACAGAATGGGGTGTTTATCTGGGAGGTTCAAAATCCCCTCTATTTCAAGATAACAGAGCACCACAACAGGGGATTCCTCATGAACCAAGACATCATCACTGCCCAGATACAGTTCAATCACAACCTGAGGAAAGCGTTGGGAATACACAAATGTTTCCTAGCCTTCCGAATCTGGATGACCTCACAGCCTCCGACTGGTCGTTTCTTAAGGGTCTTTAA |
| Protein Sequence | MQHSSPSQSHCTQVPIKVQHRLAKRRAIRRKRVDLECGCPNYVHINPHNHGFTHRGIHHCSSSTEWGVYLGGSKSPLFQDNRAPQQGIPHEPRHHHCPDTVQSQPEESVGNTQMFPSLPNLDDLTASDWSFLKGL |
| NCBI Accession | YP_009051691.1 |
|---|---|
| Location | 1598-2629 |
| Gene Name | AC1 |
| Protein Name | replication associated protein |
| Coding Region | ATGTCACGTGCCTTCCGCATAAATGCTAAGAATTACTTCCTCACTTATCCCCAATGCTCTCTAACTAAAAAAGAAGCCCTCTATCAATTAAAAAACATACACCCCCCAGTAAACAAAAAAATCATCAGGATCTGCAGAGAGCTTCACGAAAAGGGGGAACCTCATCTTCACGTGCTCATCCAGTTCGAGGGGAAGTACCAGTGCACAAATAACCGATTCTTCGACCTGGTATCGCCAACCAGGTCAGCACATTTCCATCCTAACATTCATGGAGCTAAATCAAGCTCGGACGTCAAGTCCTATCTGGAGAAGACGGAGACACCCTCGATTGGGGAGAGTTTCAGATCGATGGAAGATCTGCACGAGGGGGACAACAGTCAGCCAATGCATGCTTACGCCAAGGCAATTAACACAGGCAGTAAGTCAGAGCCTCTTAGAGTCATTAGGGAATTAGCACCAAGGGATTATGTCTTAAAATTTCCTAATCTTAATGCTAATCTAGATGGGATTTTTTCACCCCAAATGGAGGTTTATGTTTCCCCTTTTCTTTCTTCTTCCTTCGATCAAGTTCCAGAGGCCATAGAGGAATGGGCCTCTGATAATGTGATGGGTCCCGCTGCGCGGCCATTGAGACCTAAAAGTATCGTCATTGAGGGTGATAGGCGTACGGGGAAAACAATGTGGGCTAGGTCACTGGGTCCACATAATTACCTATGTGGCCATTTAGATCTGAGCCCTAAGATCTATTCAAATGATGCATGGTACAACGTCATTGATGACGTAGACCCCAACTACCTAAAGCACTTTAAAGAAATCATGGGGGCCCAGAGGGACTGGCAAAGTAACACCAAGTACGGGAAGCCAGTTCAAATTAAAGGGGGCATTCCCACGATCTTCCTATGCAATCCTGGGCCCAATTCCAGCTATAAAGAGTTCCTCGATGAGGAAAAGAACTCAGCACTGAAGTCTTGGGCTATACACAATGCAACATTCATCACCCTCACAGAGCCACTGTACTCAGGTGCCCATTAA |
| Protein Sequence | MSRAFRINAKNYFLTYPQCSLTKKEALYQLKNIHPPVNKKIIRICRELHEKGEPHLHVLIQFEGKYQCTNNRFFDLVSPTRSAHFHPNIHGAKSSSDVKSYLEKTETPSIGESFRSMEDLHEGDNSQPMHAYAKAINTGSKSEPLRVIRELAPRDYVLKFPNLNANLDGIFSPQMEVYVSPFLSSSFDQVPEAIEEWASDNVMGPAARPLRPKSIVIEGDRRTGKTMWARSLGPHNYLCGHLDLSPKIYSNDAWYNVIDDVDPNYLKHFKEIMGAQRDWQSNTKYGKPVQIKGGIPTIFLCNPGPNSSYKEFLDEEKNSALKSWAIHNATFITLTEPLYSGAH |
| NCBI Accession | YP_009051692.1 |
|---|---|
| Location | 2176-2361 |
| Gene Name | AC4 |
| Protein Name | AC4 |
| Coding Region | ATGGAGCTAAATCAAGCTCGGACGTCAAGTCCTATCTGGAGAAGACGGAGACACCCTCGATTGGGGAGAGTTTCAGATCGATGGAAGATCTGCACGAGGGGGACAACAGTCAGCCAATGCATGCTTACGCCAAGGCAATTAACACAGGCAGTAAGTCAGAGCCTCTTAGAGTCATTAGGGAATTAG |
| Protein Sequence | MELNQARTSSPIWRRRRHPRLGRVSDRWKICTRGTTVSQCMLTPRQLTQAVSQSLLESLGN |
References More References in PubMed
| 1 |
Shakir S, et al. Virol J. 2018 Aug 31;15(1):134. doi: 10.1186/s12985-018-1047-y. PMID: 30165872 |
|---|---|
| 2 |
No evidence of seed transmissibility of Tomato yellow leaf curl virus in Nicotiana benthamiana. Rosas-Díaz T, et al. J Zhejiang Univ Sci B. 2017 May;18(5):437-440. doi: 10.1631/jzus.B1600463. PMID: 28471116 |
| 3 |
Interspecies Recombination-Led Speciation of a Novel Geminivirus in Pakistan. Lal A, et al. Viruses. 2022 Sep 30;14(10):2166. doi: 10.3390/v14102166. PMID: 36298721 |
| 4 |
Diksha D, et al. Virus Genes. 2024 Oct;60(5):568-571. doi: 10.1007/s11262-024-02087-2. Epub 2024 Jun 27. PMID: 38935183 |
| 5 |
Recombinase polymerase amplification applied to plant virus detection and potential implications. Babu B, et al. Anal Biochem. 2018 Apr 1;546:72-77. doi: 10.1016/j.ab.2018.01.021. Epub 2018 Jan 31. PMID: 29408177 |
| 6 |
Gao M, et al. Mol Plant. 2025 Jun 2;18(6):1029-1046. doi: 10.1016/j.molp.2025.05.011. Epub 2025 May 26. PMID: 40420479 |
| 7 |
Mao MJ, et al. Bing Du Xue Bao. 2008 Jan;24(1):64-8. PMID: 18320825 |
| 8 |
Du Z, et al. Virol J. 2015 Oct 7;12:163. doi: 10.1186/s12985-015-0397-y. PMID: 26445958 |
| 9 |
Split-Luciferase Complementation Imaging Assay in Virus-Plant Interactions. Tan H, et al. Methods Mol Biol. 2024;2724:235-245. doi: 10.1007/978-1-0716-3485-1_17. PMID: 37987910 |
| 10 |
Huang CH, et al. Plant Dis. 2020 May;104(5):1318-1327. doi: 10.1094/PDIS-06-19-1223-RE. Epub 2020 Mar 16. PMID: 32181724 |