Melon chlorotic leaf curl virus
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_001430695.1 |
| Isolate | Guatemala |
| Release date | 2015/11/3 |
| Submitter | Brown,J.K., Idris,A.M., Rogan,D., Hussein,M.H., Palmieri,M., Mills-Lujan,K., Martin,K. |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
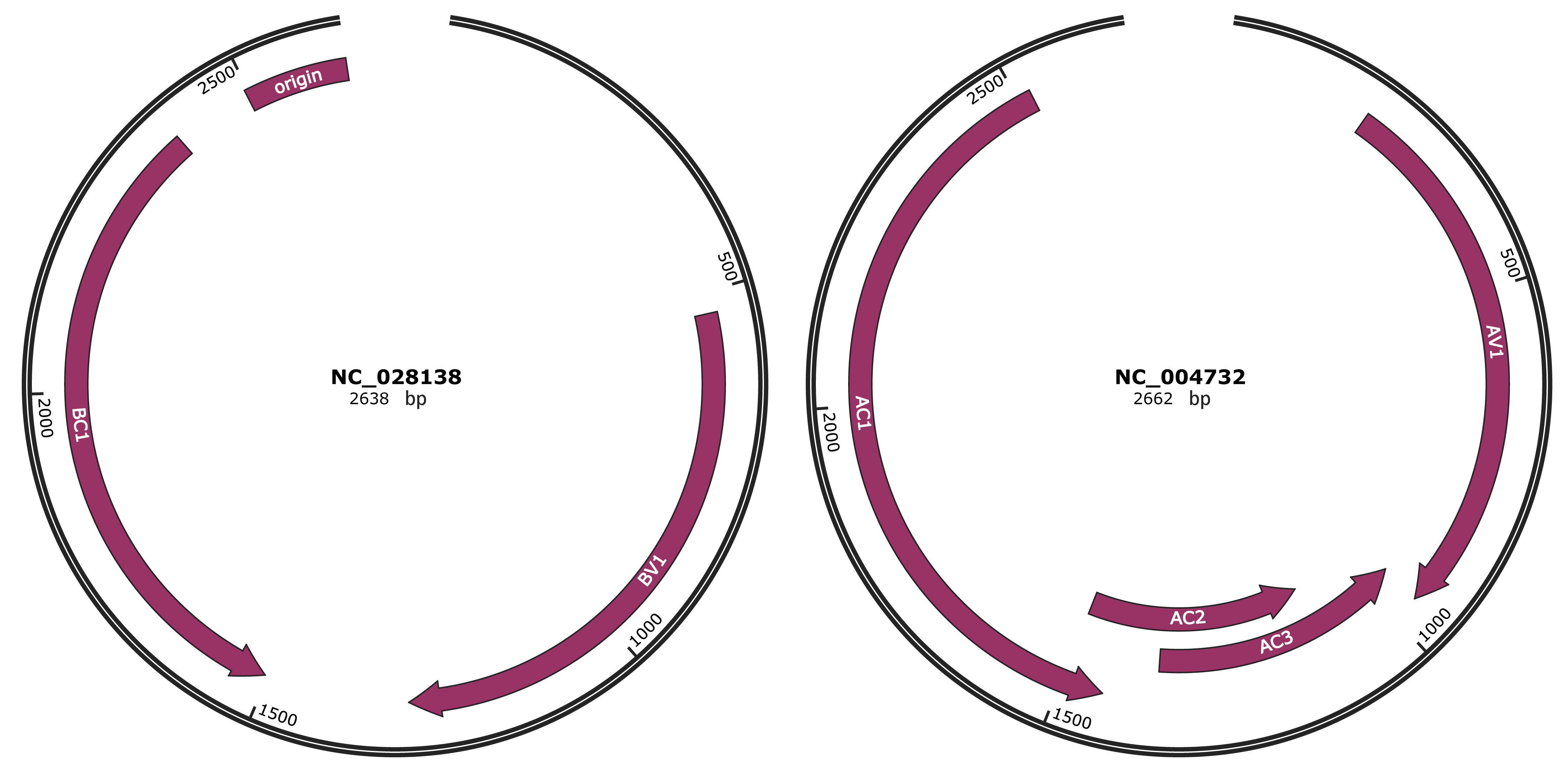
JBrowse
Genome
NC_028138
NC_004732
Gene Information
| NCBI Accession | YP_009175072.1 |
|---|---|
| Location | 530-1300 |
| Gene Name | BV1 |
| Protein Name | nuclear shuttle protein |
| Coding Region | ATGTATTCGGCGAACTATAGACGTACTCGTGGGACTACCCAACGTGGTTCATATTCACGACGTCCAATATTCAAGCGTCCACATTATTTGTCTCGGTCAGATGATAAGCGTCGCACTAATTCGACTATTGTGCCTCATGCCGACACTAAGATGTTAAAACAACGGTTGCATGAGGATCAATTTGGCCCAGATTATGTTTTGTGTCATAATAGTGCGATGTCCACGTTTATTACTCTTCCTAGTCTTGGTAAGCACGAGCCCAATCGTTCAAGGTCGTTTATAAAGTTACAACGACTCCGTTACAAGGGAACTCTGAAAATTGAACGTGGACCTGGTGACACGTTCATGGAAGGTCCCACGTCAAAGGTAGAAGGCGTCTTTTCTATGGTAGTTGTGGTTGATCGTAAACCACATGTTAACCCATCTGGACGCTTACATTCCTTTGATGAGTTATTCGGCGCGCGTATCCATAGTCACGGTAATTTAGCCATAGTCCCGGCATTGAAGGAGCGTTTTTACATTCGCCACGTATTGAAACGAGTATTATCCGTCGAGAAAGACACGTTGATGGTTGATCTACATGGAACAACCGCATTATCTAATAGGCGATTCAATTGTTGGTCCTCGTTTAATGATCTTGAACGCGATTCATGTAACGGCGTTTACGCTAATATAAACAAGAACGCTCTTTTAGTTTATTATTGTTGGATGTCGGATATGCCTTCGAAGGCATCGACATATGTAACGTTTGATCTCGACTATGTCGGATAA |
| Protein Sequence | MYSANYRRTRGTTQRGSYSRRPIFKRPHYLSRSDDKRRTNSTIVPHADTKMLKQRLHEDQFGPDYVLCHNSAMSTFITLPSLGKHEPNRSRSFIKLQRLRYKGTLKIERGPGDTFMEGPTSKVEGVFSMVVVVDRKPHVNPSGRLHSFDELFGARIHSHGNLAIVPALKERFYIRHVLKRVLSVEKDTLMVDLHGTTALSNRRFNCWSSFNDLERDSCNGVYANINKNALLVYYCWMSDMPSKASTYVTFDLDYVG |
| NCBI Accession | YP_009175073.1 |
|---|---|
| Location | 1505-2386 |
| Gene Name | BC1 |
| Protein Name | movement protein |
| Coding Region | ATGGGTTCTCAATTAGCTCCTCCACCGAATGCGTTCAATTATATAGAGTCTAGTCGGGATGAATTTCAGTTATCGCACGACTTAACGGAAATTATTCTGCAATTTCCGTCGACTACTTCCCAATTAACTGCAAGACTTAGTCGGAGTTGTATGAAGATCGACCATTGTGTCATAGAATATAGGCAACAAGTACCTATCAACGCATCTGGAACAGTAATAGTGGAGATTCACGATAAACGAATGACCGACAATGAGTCCTTACAAGCCTCGTGGACGTTTCCGATCAGATGTAACATAGATCTCCACTATTTCTCGTCATCATTCTTCTCTCTCAAAGATCCCATTCCTTGGAAGTTATATTATCGGGTAACAGACTCGAACGTGCATCAGATGACTCATTTCGCTAAATTCAAAGGTAAATTGAAGTTATCGTCGGCGAAACATTCCGTCGATATTCCCTTCCGGGCACCAACAGTCAAAATCCTTACAAAGCAATTTAGCCAAAAGGACGTCGATTTCTGGCACGTTGGGTACGGCAAATGGGAGAGAAGACTGGTCAAATCCGCATCGATGTCAAGATATGGGCTCAGAGGCCCAATTGAATTAAATCCAGGCGAATCTTGGGCCACTAGAAGTGCTCTGGGTACGAGCCCACTAAATGCGGACTTGGATACAACAGATGAGATGCTTCCCTATCGCGAGCTTAACAGGTTGGGTACATCTATATTAGACCCTGGAGAATCGGCGTCAATGGTCGGAATACAACGGTCGCAATCAAACATTACCATGTCAATGGCACAACTGAACGAATTAGTTAGATCCACCGTTCAGGAGTGCATCAAGACCAGTTGTATTCCCTCAACTCCAAAATCCTTAGGATAA |
| Protein Sequence | MGSQLAPPPNAFNYIESSRDEFQLSHDLTEIILQFPSTTSQLTARLSRSCMKIDHCVIEYRQQVPINASGTVIVEIHDKRMTDNESLQASWTFPIRCNIDLHYFSSSFFSLKDPIPWKLYYRVTDSNVHQMTHFAKFKGKLKLSSAKHSVDIPFRAPTVKILTKQFSQKDVDFWHVGYGKWERRLVKSASMSRYGLRGPIELNPGESWATRSALGTSPLNADLDTTDEMLPYRELNRLGTSILDPGESASMVGIQRSQSNITMSMAQLNELVRSTVQECIKTSCIPSTPKSLG |
| NCBI Accession | NP_835272.1 |
|---|---|
| Location | 206-961 |
| Gene Name | AV1 |
| Protein Name | coat protein |
| Coding Region | ATGGTTAAGAGGGATGCCCCATGGCGTTTAATGGCGGGGACCTCCAAGGTTTCCCGCTCTGCTAATTATTCGCCTCGTGGAGGTATGGGCCCTAAATTCAATAAGGCCGCTGCATGGGTTAATAGGCCCATGTACAGGAAGCCCAGAATTTATCGCACGTTACGAGGTCCTGACATCCCCAAAGGATGTGAAGGACCATGCAAGGTTCAATCATTTGAGCAGCGGCATGATATCTCTCATATTGGTAAGGTTATGTGTATTTCTGACGTGACGCGTGGCAATGGTATTACTCACCGCGTCGGCAAGCGTTTTTGTGTTAAGTCTGTATATATTTTAGGCAAGATATGGATGGACGAAAACATCAAGCTGAAGAATCACACAAACAGTGTGATGTTTTGGCTAGTTCGAGATCGGAGACCCTATGGCACTCCTATGGATTTCGGCCAAGTATTCAACATGTTTGACAACGAGCCTAGCACTGCAACCGTTAAGAACGATCTACGAGATCGTTATCAAGTTTTGCACAGGTTTTACGCGAAAGTCACTGGTGGTCAGTATGCTAGCAACGAACAAGCCTTGGTGAGGCGTTTTTGGAAGGTCAACAATTATGTTGTCTACAATCACCAGGAAGCAGCAAAATACGAGAATCATACGGAGAACGCTTTATTATTGTATATGGCATGTACTCATGCATCTAACCCTGTGTACGCGACATTGAAAATTCGGATCTATTTTTATGATTCGATAACAAATTAA |
| Protein Sequence | MVKRDAPWRLMAGTSKVSRSANYSPRGGMGPKFNKAAAWVNRPMYRKPRIYRTLRGPDIPKGCEGPCKVQSFEQRHDISHIGKVMCISDVTRGNGITHRVGKRFCVKSVYILGKIWMDENIKLKNHTNSVMFWLVRDRRPYGTPMDFGQVFNMFDNEPSTATVKNDLRDRYQVLHRFYAKVTGGQYASNEQALVRRFWKVNNYVVYNHQEAAKYENHTENALLLYMACTHASNPVYATLKIRIYFYDSITN |
| NCBI Accession | NP_835273.1 |
|---|---|
| Location | 958-1362 |
| Gene Name | AC3 |
| Protein Name | replicase-associated protein |
| Coding Region | ATGGTAATGGATTTACGCACTTGGGAAGACATCACTGTGTATCAGGCGGAGAATTCCGTTTTTATCTGGGAAGTTCCAAATCCCCTCTATTTCAAGATATACAAAGTGGAGGATCCACTGTATACGCACACAAGAATATACCACATACAAATCCGATTCAACCACAACCTGCGGAGAGCGCTAAATCTTCACAAAGCATACCTGAACTTCCAAGTCTGGACGGAATCGATTCGAGCTTCTGGGACGACTTATTTAAATAGATTTAAACATTTAGTTATGTTGTATTTAGATAGATTAGGGGTTATCGGTCTCAATAACGTCATACGTGCTGTATCATGGGCGACAGACCGTTCATATGTAAATTATGTAATCGAGAATCATATAATAAAATTCAACGTTTATTAA |
| Protein Sequence | MVMDLRTWEDITVYQAENSVFIWEVPNPLYFKIYKVEDPLYTHTRIYHIQIRFNHNLRRALNLHKAYLNFQVWTESIRASGTTYLNRFKHLVMLYLDRLGVIGLNNVIRAVSWATDRSYVNYVIENHIIKFNVY |
| NCBI Accession | NP_835274.1 |
|---|---|
| Location | 1103-1498 |
| Gene Name | AC2 |
| Protein Name | transactivation protein |
| Coding Region | ATGCCCAATTCATCTTCCTCGACTGCCCCCTCTATCAAAGCACAACACAAGATCGCTAAAACAAGAGCTATCCGTCGTAGACGCATCGACCTGAACTGCGGCTGTTCCATATTCCTCCATCTTAGCTGCCAAAAAAATGGTAATGGATTTACGCACTTGGGAAGACATCACTGTGTATCAGGCGGAGAATTCCGTTTTTATCTGGGAAGTTCCAAATCCCCTCTATTTCAAGATATACAAAGTGGAGGATCCACTGTATACGCACACAAGAATATACCACATACAAATCCGATTCAACCACAACCTGCGGAGAGCGCTAAATCTTCACAAAGCATACCTGAACTTCCAAGTCTGGACGGAATCGATTCGAGCTTCTGGGACGACTTATTTAAATAG |
| Protein Sequence | MPNSSSSTAPSIKAQHKIAKTRAIRRRRIDLNCGCSIFLHLSCQKNGNGFTHLGRHHCVSGGEFRFYLGSSKSPLFQDIQSGGSTVYAHKNIPHTNPIQPQPAESAKSSQSIPELPSLDGIDSSFWDDLFK |
| NCBI Accession | NP_835275.1 |
|---|---|
| Location | 1440-2519 |
| Gene Name | AC1 |
| Protein Name | replication-associated protein |
| Coding Region | ATGCCACAGAACCCTAAATCGTTCCGCTTATCTGCCAGAAATATTTTCCTAACATATCCGAAGTGCGATGTTCCCAAAGATGAAGTTCTTGAAATACTGCAAGGTCTATCCTGGTCAGTCGTCAAACCGACATATATCCGTGTCGCACGAGAGGAACACGCAGATGGGTTCCCGCACTTACACTGCCTCGTTCAACTTTCAGGCAAGTCAAACATCAAGGATGCTGGATTTTTCGACATTACTCACCCAAGAAGGTCTGCCCGATTTCACCCTAATATTCAAGCTGCCAAAGACACCAATGCCGTCAAGAATTACATCACCAAAGAAGGTGATTATTGTGAATTCGGGCAATATAAGGTGTCTGGGGGTTCAAAGTCCAATAAAGACGACGTTTATCACAACGCCGTCAATGCAGCAAGTGCGGGAGAGGCTCTCGACATTATAAAAGCTGGCGATCCAAAGACGTTTATAGTGAGTTACCATAACATAACGGCTAACGTCGATCGCCTATTTCATAAACCTCCAATGCCATGGGTTCCCCCATATGAACTTTCTTCGTTTACAAATGTTCCAGAAGAGTTAAAAGATTGGGCTGATCATTATTTTAATGAATGTTCAGCTGCGCGGCCATTTTTAATAAAAGAATCAAATAACGTGTGTAATAGACCTATTAGTGTAATAATTGAGGGTGATTCTCGAACGGGCAAAACAATGTGGGCTCGGTCTTTAGGCCCACATAACTATTTGAGTGGACACCTTGATTTCAATTCTAGGGTTTTCTCAAATGACGTCAAGTATAACGTCATTGATGACGTCGCTCCGCATTATTTAAAGCTAAAGCACTGGAAAGAATTGATTGGGGCCCAAAGGGACTGGCAGTCCAACTGTAAATATGGAAAGCCGGTTCAAATTAAAGGTGGAATCCCATCAATAGTGCTTTGCAATCCCGGAGAGGGGGCCAGCTATAAAGAGTTTCTCGACAAAGCTGAAAATGCAGCACTAAGAGAGTGGACACTAAAAAATGCCCAATTCATCTTCCTCGACTGCCCCCTCTATCAAAGCACAACACAAGATCGCTAA |
| Protein Sequence | MPQNPKSFRLSARNIFLTYPKCDVPKDEVLEILQGLSWSVVKPTYIRVAREEHADGFPHLHCLVQLSGKSNIKDAGFFDITHPRRSARFHPNIQAAKDTNAVKNYITKEGDYCEFGQYKVSGGSKSNKDDVYHNAVNAASAGEALDIIKAGDPKTFIVSYHNITANVDRLFHKPPMPWVPPYELSSFTNVPEELKDWADHYFNECSAARPFLIKESNNVCNRPISVIIEGDSRTGKTMWARSLGPHNYLSGHLDFNSRVFSNDVKYNVIDDVAPHYLKLKHWKELIGAQRDWQSNCKYGKPVQIKGGIPSIVLCNPGEGASYKEFLDKAENAALREWTLKNAQFIFLDCPLYQSTTQDR |
References More References in PubMed
| 1 |
Maliano MR, et al. Plant Dis. 2021 Oct;105(10):3162-3170. doi: 10.1094/PDIS-08-20-1759-RE. Epub 2021 Nov 7. PMID: 33591835 |
|---|---|
| 2 |
Resistance Against Melon Chlorotic Mosaic Virus and Tomato Leaf Curl New Delhi Virus in Melon. Romay G, et al. Plant Dis. 2019 Nov;103(11):2913-2919. doi: 10.1094/PDIS-02-19-0298-RE. Epub 2019 Aug 20. PMID: 31436474 |
| 3 |
García-Rodríguez DA, et al. PeerJ. 2023 Mar 22;11:e15047. doi: 10.7717/peerj.15047. eCollection 2023. PMID: 36974135 |
| 4 |
Brown JK, et al. Plant Dis. 2001 Sep;85(9):1027. doi: 10.1094/PDIS.2001.85.9.1027C. PMID: 30823088 |
| 5 |
Esmaeili M, et al. Virus Genes. 2015 Dec;51(3):408-16. doi: 10.1007/s11262-015-1250-5. Epub 2015 Oct 3. PMID: 26433951 |
| 6 |
Brown JK, et al. Virus Res. 2011 Jun;158(1-2):257-62. doi: 10.1016/j.virusres.2011.03.002. Epub 2011 Mar 17. PMID: 21420452 |
| 7 |
Fontenele RS, et al. Viruses. 2021 Apr 30;13(5):810. doi: 10.3390/v13050810. PMID: 33946382 |
| 8 |
Idris AM, et al. J Virol. 2008 Feb;82(4):1959-67. doi: 10.1128/JVI.01992-07. Epub 2007 Dec 5. PMID: 18057231 |
| 9 |
Sufrin-Ringwald T, et al. Phytopathology. 2011 Feb;101(2):281-9. doi: 10.1094/PHYTO-06-10-0159. PMID: 21219130 |
| 10 |
Disease complex associated with begomoviruses infecting squash and cucumber in Saudi Arabia. Amer MA, et al. Cell Mol Biol (Noisy-le-grand). 2024 Nov 27;70(11):58-63. doi: 10.14715/cmb/2024.70.11.8. PMID: 39707780 |