Macroptilium yellow spot virus
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_000895435.1 |
| Isolate | Brazil |
| Release date | 2015/2/22 |
| Submitter | Silva,S.J.C., Castillo-Urquiza,G.P., Hora,B.T. Jr., Assuncao,I.P., Lima,G.S.A., Pio-Ribeiro,G., Mizubuti,E.S.G., Zerbini,F.M. |
| Host | |
| Vector | |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
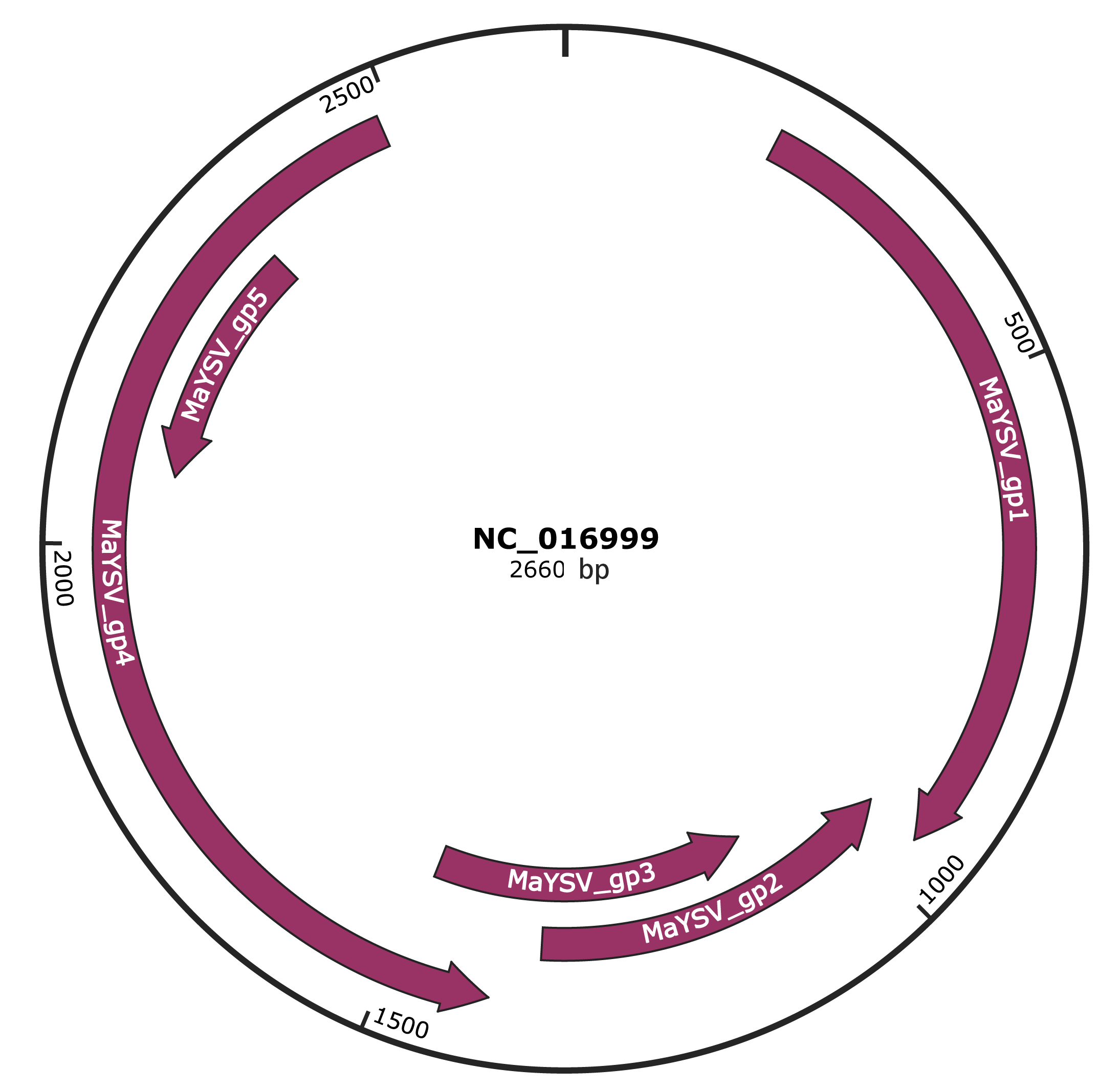
JBrowse
Genome
NC_016999
Gene Information
| NCBI Accession | YP_005352895.1 |
|---|---|
| Location | 204-959 |
| Protein Name | coat protein |
| Coding Region | ATGGTTAAGCGGGACGCCCCATGGCGCCACATGGCAGGAACCTCAAAGGTTTCCCGCAATAACAACTATTCCCCTCGTGGAGGTGGAGGTCCAAAAAACAACAGGGCCTCTGAATGGGTTAATAGGCCCATGTACAGAAAGCCCAGGATATATCGTATGTACAGAACCCCCGATGTGCCTAGGGGGTGTGAAGGCCCATGTAAGGTTCAGTCATTTGAACAGCGACACGATATATCCCATGTTGGTAAGGTTATATGCATGTCTGACGTGACAAGAGGTAATGGTATTACCCATCGTGTAGGAAAACGTTTTTGTGTGAAGTCTGTTTATATTCTAGGTAAGATATGGATGGACGAGAACATCAAGTTGAAGAATCACACGAACAGTGTGATGTTTTGGTTGGTCAGAGACAGAAGACCCTATGGCTCTCCCATGGACTTTGGCCAAGTGTTCAACATGTTTGACAACGAGCCCAGTACTGCTACGGTTAAGAACGATCTTCGTGATCGTTTTCAAGTCATGCATCGATTCTATGCCAAAGTCACTGGTGGACAGTATGCGAGCAACGAGCAAGCAATTGTCAGGCGTTTTTGGAAGGTCAACAATCATGTTGTCTACAACCACCAGGAAGCAGGGAAATACGAGAATCATACGGAGAACGCTTTACTATTGTATATGGCATGTACTCATGCCTCTAACCCCGTGTATGCAACCTTAAAAATTCGGATCTATTTTTATGATTCGATAACCAATTAA |
| Protein Sequence | MVKRDAPWRHMAGTSKVSRNNNYSPRGGGGPKNNRASEWVNRPMYRKPRIYRMYRTPDVPRGCEGPCKVQSFEQRHDISHVGKVICMSDVTRGNGITHRVGKRFCVKSVYILGKIWMDENIKLKNHTNSVMFWLVRDRRPYGSPMDFGQVFNMFDNEPSTATVKNDLRDRFQVMHRFYAKVTGGQYASNEQAIVRRFWKVNNHVVYNHQEAGKYENHTENALLLYMACTHASNPVYATLKIRIYFYDSITN |
| NCBI Accession | YP_005352896.1 |
|---|---|
| Location | 956-1354 |
| Protein Name | replication-enhancer protein |
| Coding Region | ATGGATTCACGCACAGGGGAACTCATCACTGCACATCAGGCAGAGAGTGGCGTTTATATCTGGGAGATATCAAATCCCCTTTATTTCAAGATCAAACAGGTGGAGGATCCACTATACACGACAACCAGAATATATCACGTCCAGATCAGGTTCAACCACAACCTCAGGAGAGCATTGGCTCTCCACAAGGCATATCTCAATTTCCAAGTCTGGACGATATCCCTGAGAGCTTCTGGGACGACATATTTAGTTAGGTTTAAATACTTAGTCATGTTGTACTTAGGACGAATAGGTGTTGTATCGCTTAACAATGTAATCAGAGCTGTTCGATTTGCAACAGACAAATCATATGTAAATTATGTACTTGAGAATCATGAAATAAAATTCAAATTTTATTAA |
| Protein Sequence | MDSRTGELITAHQAESGVYIWEISNPLYFKIKQVEDPLYTTTRIYHVQIRFNHNLRRALALHKAYLNFQVWTISLRASGTTYLVRFKYLVMLYLGRIGVVSLNNVIRAVRFATDKSYVNYVLENHEIKFKFY |
| NCBI Accession | YP_005352897.1 |
|---|---|
| Location | 1101-1490 |
| Protein Name | transactivating protein |
| Coding Region | ATGCGCAATTCGTCTTCCTCAACTCCCCCCTCTATCAAGGTTCAACATACAGCGGCCAAGAAGAGAGCAATCAGAAGAAGACGAATTGACATAGATTGCGGGTGCTCCATATACGTCCATATCAACTGCAGAGGACATGGATTCACGCACAGGGGAACTCATCACTGCACATCAGGCAGAGAGTGGCGTTTATATCTGGGAGATATCAAATCCCCTTTATTTCAAGATCAAACAGGTGGAGGATCCACTATACACGACAACCAGAATATATCACGTCCAGATCAGGTTCAACCACAACCTCAGGAGAGCATTGGCTCTCCACAAGGCATATCTCAATTTCCAAGTCTGGACGATATCCCTGAGAGCTTCTGGGACGACATATTTAGTTAG |
| Protein Sequence | MRNSSSSTPPSIKVQHTAAKKRAIRRRRIDIDCGCSIYVHINCRGHGFTHRGTHHCTSGREWRLYLGDIKSPLFQDQTGGGSTIHDNQNISRPDQVQPQPQESIGSPQGISQFPSLDDIPESFWDDIFS |
| NCBI Accession | YP_005352898.1 |
|---|---|
| Location | 1402-2487 |
| Protein Name | replication-associated protein |
| Coding Region | ATGCCACCGCCCAGACGTTTTAAAATTCAGTCCAAAAATTATTTCCTTACTTATCCCAAGTGCTCTATCTCCAAAGAAGAGGCACTTGCTCAACTTCTAGCATTAGATACACCTACAAACAAGAAATATATCCGTGTATGCAGAGAATTACACGGAAATGGGGAGCCTCATCTCCATGCTTTGCTGCAATTCGAAGGTAAGTTCACATGCACGAATTGCAGATTCTTCGACTTGCGACATCCCAGTCATTCAAATGTTTGTCATGGAAAATACGAATCATGCAAGTCATCCTCCGACGCCAAATCATATATTGAGAAGGACGGAGATTACGTCGAATGGGGTGATTTTCAGATCGACGGACGATCTGCTCGAGGAGGTCAACAGACAGTTAACGATACATATGCAAAGGCGTTAAATGCTTCTTCAGCAGAAGAAGCACTTCAAATAATAAAGGAGGAACAACCACAGCACTTCTTCCTCCAACATCACAATCTTGTGGCTAACGCATATCGTATATTTCAAAAGGCTCCGGAGCCATGGACTCCTCCGTTTCCACTCTCTTCTTTCATTAATGTCCCAGAAGAGATGAAAGCTTGGGCAGATGACTATTTCGGAAGAAGTGCCGCTGCGCGGCCGGAGAGACCAATTAGTATAATCATCGAGGGTGACTCTCGAACGGGCAAGACCATGTGGGCACGTGCATTGGGGTCCCATAATTACTTGAGTGGTCACCTCGATTTCAATTCAAAGGTCTATTCAAATGATGTGCAGTACAACGTGATAGATGACATCGCACCGCAATATCTAAAGTTAAAGCACTGGAAAGAATTGATAGGTGCTCAAAAGGACTGGCAATCAAATTGCAAATACGGGAAGCCAGTTCAAATTAAAGGTGGAATCCCATGCATTGTGCTTTGCAATCCTGGCGAGGGCGCCAGCTATAAATCGTTCCTCGACAAAGAAGAAAACGCATCACTTAGACAGTGGACGATACACAATGCGCAATTCGTCTTCCTCAACTCCCCCCTCTATCAAGGTTCAACATACAGCGGCCAAGAAGAGAGCAATCAGAAGAAGACGAATTGA |
| Protein Sequence | MPPPRRFKIQSKNYFLTYPKCSISKEEALAQLLALDTPTNKKYIRVCRELHGNGEPHLHALLQFEGKFTCTNCRFFDLRHPSHSNVCHGKYESCKSSSDAKSYIEKDGDYVEWGDFQIDGRSARGGQQTVNDTYAKALNASSAEEALQIIKEEQPQHFFLQHHNLVANAYRIFQKAPEPWTPPFPLSSFINVPEEMKAWADDYFGRSAAARPERPISIIIEGDSRTGKTMWARALGSHNYLSGHLDFNSKVYSNDVQYNVIDDIAPQYLKLKHWKELIGAQKDWQSNCKYGKPVQIKGGIPCIVLCNPGEGASYKSFLDKEENASLRQWTIHNAQFVFLNSPLYQGSTYSGQEESNQKKTN |
| NCBI Accession | YP_005352899.1 |
|---|---|
| Location | 2073-2330 |
| Protein Name | AC4 protein |
| Coding Region | ATGGGGAGCCTCATCTCCATGCTTTGCTGCAATTCGAAGGTAAGTTCACATGCACGAATTGCAGATTCTTCGACTTGCGACATCCCAGTCATTCAAATGTTTGTCATGGAAAATACGAATCATGCAAGTCATCCTCCGACGCCAAATCATATATTGAGAAGGACGGAGATTACGTCGAATGGGGTGATTTTCAGATCGACGGACGATCTGCTCGAGGAGGTCAACAGACAGTTAACGATACATATGCAAAGGCGTTAA |
| Protein Sequence | MGSLISMLCCNSKVSSHARIADSSTCDIPVIQMFVMENTNHASHPPTPNHILRRTEITSNGVIFRSTDDLLEEVNRQLTIHMQRR |
References More References in PubMed
| 1 |
Collins A, et al. Virus Res. 2010 Jun;150(1-2):148-52. doi: 10.1016/j.virusres.2010.03.008. Epub 2010 Mar 25. PMID: 20347895 |
|---|---|
| 2 |
Contrasting genetic structure between two begomoviruses infecting the same leguminous hosts. Sobrinho RR, et al. J Gen Virol. 2014 Nov;95(Pt 11):2540-2552. doi: 10.1099/vir.0.067009-0. Epub 2014 Jul 15. PMID: 25028472 |
| 3 |
Macedo MA, et al. Arch Virol. 2018 Mar;163(3):737-743. doi: 10.1007/s00705-017-3662-0. Epub 2017 Dec 9. PMID: 29224131 |
| 4 |
Lett JM, et al. Arch Virol. 2015 Nov;160(11):2887-90. doi: 10.1007/s00705-015-2558-0. Epub 2015 Aug 9. PMID: 26255054 |
| 5 |
Lima ATM, et al. J Gen Virol. 2013 Feb;94(Pt 2):418-431. doi: 10.1099/vir.0.047241-0. Epub 2012 Nov 7. PMID: 23136367 |