Macroptilium yellow mosaic virus
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_000848005.1 |
| Isolate | Jamaica |
| Release date | 2015/2/12 |
| Submitter | Amarakoon,I.I., Roye,M.E., Briddon,R.W., Bedford,I.D., Stanley,J. |
| Host | |
| Vector | |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
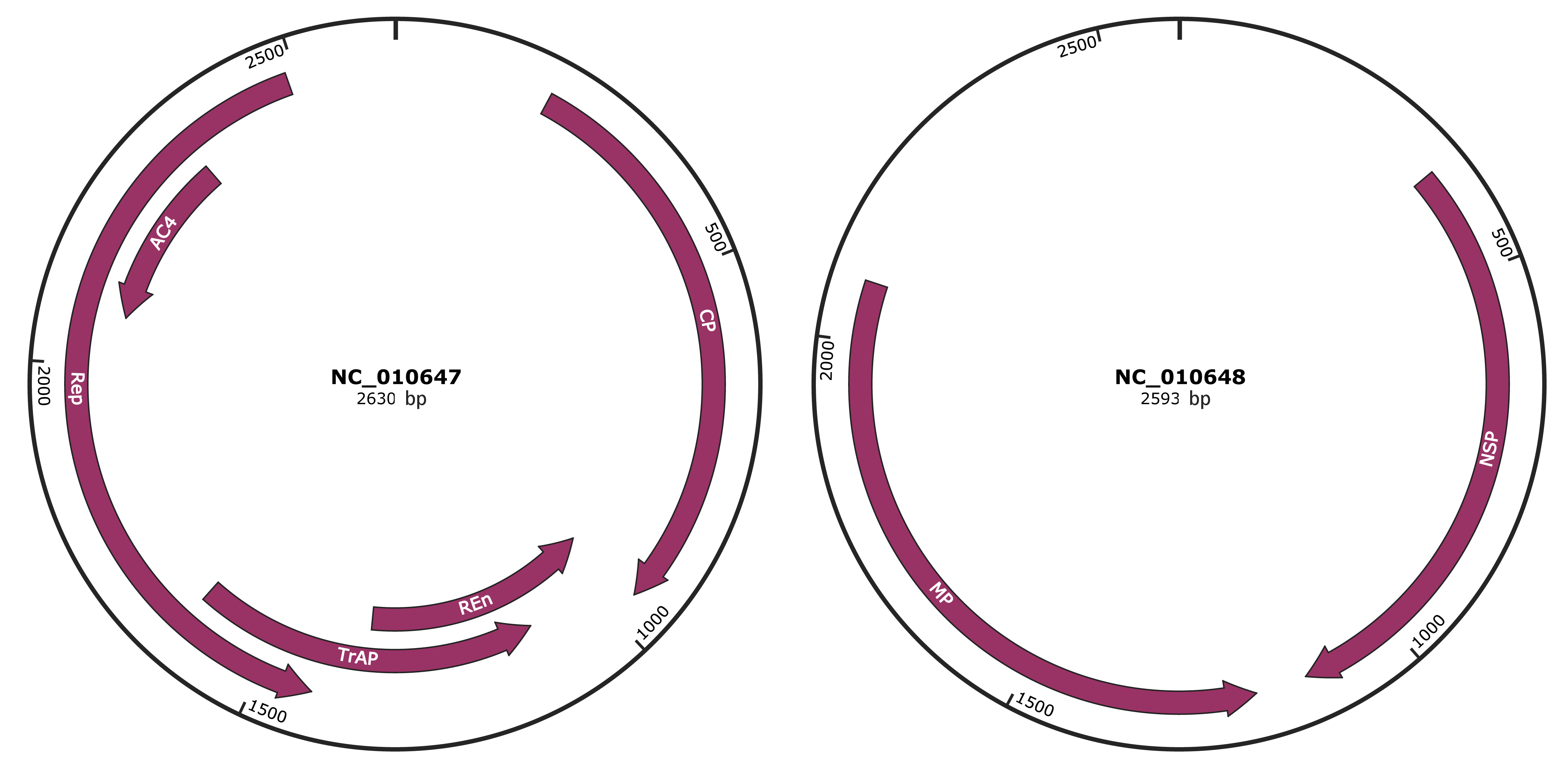
JBrowse
Genome
NC_010647
NC_010648
Gene Information
| NCBI Accession | YP_001876449.1 |
|---|---|
| Location | 208-960 |
| Gene Name | CP |
| Protein Name | coat protein |
| Coding Region | ATGCCCAAGAGGGATGCCCCGTGGCGTTTGATGGCGGGAACCTCAAAGGTTTCCCGCACTGGCAATTATTCTCGAAATGGTGGTTTGGGCCAATCCTCCAACAAAAACGCTTGGGTGAACAGGCCCATGTACAGGAAGCCCAGGATTTATCGGATGTACAGATCATCCGCTGTGCCCAAGGGATGTGAAGGACCTTGCAAGGTCCAATCCTATGAACAACGACATGATATATCTCATGTTGGTAAGGTGATATGTATTTCTGACATTACTCGTGGTAATGGTATTACTCATCGTGTAGGGAAGCGTTTTTGTGTGAAATCTGTGTATATTTTAGGGAAGATATGGATGGATGAGAACATCAAGCTCAAGAACCACACAAACAGCGTTATTTTTTGGTTGGTGCGTGATCGTCGTCCCTATGGAACACCTATGGACTTTGGACAGGTGTTCAACATGTTTGATAATGAGCCCAGTACAGCTACTGTGAAGAACGATCTTCGTGATCGTTTCCGAGTGATACACAGATTTCATGCTAAGGTGACTGGTGGCCAATATGCAAGTAATGAACAGGCATTGGTAAGGCGATTTTGGAAGGTGAACAACCATGTCGTGTACAACCACCAGGAATCCGGAAAATACGAGAATCATACGGAGAACGCCTTATTATTGTATATGGCATGTACACATGCCTCAAACCCTGTATATGCAACATTGAAAATTCGGATCTATTTCTATGATTCGATAACAAATTAA |
| Protein Sequence | MPKRDAPWRLMAGTSKVSRTGNYSRNGGLGQSSNKNAWVNRPMYRKPRIYRMYRSSAVPKGCEGPCKVQSYEQRHDISHVGKVICISDITRGNGITHRVGKRFCVKSVYILGKIWMDENIKLKNHTNSVIFWLVRDRRPYGTPMDFGQVFNMFDNEPSTATVKNDLRDRFRVIHRFHAKVTGGQYASNEQALVRRFWKVNNHVVYNHQESGKYENHTENALLLYMACTHASNPVYATLKIRIYFYDSITN |
| NCBI Accession | YP_001876450.1 |
|---|---|
| Location | 957-1355 |
| Gene Name | REn |
| Protein Name | replication enhancer protein |
| Coding Region | ATGGATTCACGCACAGGGGAGAACATCACTGCGCGTCAGGCAGAGAATTCCGTTTTTATCTGGGAGGTTCCAAATCCCCCCTATTTCAAGATCATCATGGTGGAGGATCCACCATTCACTCGAACAAGGATCTACCACATACAAGTCAGAGCCAACCACAACGTGCGGAAAGTGTTGGGTCTCCACAAAGCCTTCTTCAACTTCCAAGTCTGGACGACATTAACGACAGCTTCTGGGACGACATATTTAAATAGATTTAAATATCTTGTTCATATGTATCCTGATCAAATAGGGCTTGTCGGAATAAACAATGTAATTAGAGCTGTTCGTTTTGCCACGGACAAATCATATGTAAATTATGTGCTCGAAAGTCATGAAATAAAATTCAAATTTTATTAA |
| Protein Sequence | MDSRTGENITARQAENSVFIWEVPNPPYFKIIMVEDPPFTRTRIYHIQVRANHNVRKVLGLHKAFFNFQVWTTLTTASGTTYLNRFKYLVHMYPDQIGLVGINNVIRAVRFATDKSYVNYVLESHEIKFKFY |
| NCBI Accession | YP_001876451.1 |
|---|---|
| Location | 1102-1620 |
| Gene Name | TrAP |
| Protein Name | transcription activator protein |
| Coding Region | ATGGAAAGCCGGTTCAAATTAAAGGAGGTATCCCTTCAATCGTGCTGTGCAATCCTGGTGAGGGTGCCAGTTATAAAGACTTCCTCGGTAAAGAAGAAAACCGAGCTTTACACAACTGGACCATTCATAATGCGCTATTCATCACCTTCACAGCCCCCCTCTATCAAAGCACAGCATCGCCTTGCCAAACGTAGGGCCATCCGTCGACGACGCTTAGACTTGAACTGCGGCTGTTCCATATTTCTCCATATCAATTGCCACGACCATGGATTCACGCACAGGGGAGAACATCACTGCGCGTCAGGCAGAGAATTCCGTTTTTATCTGGGAGGTTCCAAATCCCCCCTATTTCAAGATCATCATGGTGGAGGATCCACCATTCACTCGAACAAGGATCTACCACATACAAGTCAGAGCCAACCACAACGTGCGGAAAGTGTTGGGTCTCCACAAAGCCTTCTTCAACTTCCAAGTCTGGACGACATTAACGACAGCTTCTGGGACGACATATTTAAATAG |
| Protein Sequence | MESRFKLKEVSLQSCCAILVRVPVIKTSSVKKKTELYTTGPFIMRYSSPSQPPSIKAQHRLAKRRAIRRRRLDLNCGCSIFLHINCHDHGFTHRGEHHCASGREFRFYLGGSKSPLFQDHHGGGSTIHSNKDLPHTSQSQPQRAESVGSPQSLLQLPSLDDINDSFWDDIFK |
| NCBI Accession | YP_001876452.1 |
|---|---|
| Location | 1427-2488 |
| Gene Name | Rep |
| Protein Name | replication-associated protein |
| Coding Region | ATGCCACCACCAAAGAGATTTAGAGTGAGCGCAAAAAACTATTTCCTCACATATCCTCAGTGTTCTCTTTCAAAAGAAGAGGCTCTTTCCCAATTACAAAATATTTTCACTGCCACCAACAAGAAGTTCATCAAGATATGTGAGGAATTACACGAGGATGGGCAGCCTCATCTCCATGTGCTTATCCAGTTCGAAGGAAAATTCGTCCGCACGAATAACAGATTATTCGACCTGGTCTCCCCATCCCGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAATCGAGCTCCGACGTCAAGTCCTATCTCGACAAGGATGGAGTCACAGTCGAATGGGGAGAATTCCAGGTCGACGGTCGAAGTGCTAGAGGAGGCTGCCAGAGTGCTAACGACTCATATGCCAAGGCATTGAATGCCTCATCAGTCGAACAGGCACTTCAAATATTAAAGGAGGAGCAGCCCAAGGATTATCTTCTCCAACATCATAACATCCGTTCTAACCTAGAACGGATCTTTACCAAGGTTCCGGAACCATGGGTTCCTCCATTTCACCTCTCCTCCTTTACTCACGTTCCGGCTGAGATGCAAGCGTGGGTCGATGGTTATTTCGGAAGGGGTGCCGCTGCGCGGCCGGAGAGATATATGAGTATCATCGTCGAGGGTGACTCAAGAACAGGCAAAACAATGTGGGCTCGTTCATTGGGACCACACAATTATCTCATGGGTCACCTCGACTTCAATTCGCGTGTCTATTCCAATGCCGCTGAGTACAACGTCATTGATGACATAAGCCCCAATTATTTGAAATTAAAGCATTGGAAAGAGTTAATTGGGGCTCAAAGGGACTGGCAGTCAAACTGTAAATATGGAAAGCCGGTTCAAATTAAAGGAGGTATCCCTTCAATCGTGCTGTGCAATCCTGGTGAGGGTGCCAGTTATAAAGACTTCCTCGGTAAAGAAGAAAACCGAGCTTTACACAACTGGACCATTCATAATGCGCTATTCATCACCTTCACAGCCCCCCTCTATCAAAGCACAGCATCGCCTTGCCAAACGTAG |
| Protein Sequence | MPPPKRFRVSAKNYFLTYPQCSLSKEEALSQLQNIFTATNKKFIKICEELHEDGQPHLHVLIQFEGKFVRTNNRLFDLVSPSRSAHFHPNIQGAKSSSDVKSYLDKDGVTVEWGEFQVDGRSARGGCQSANDSYAKALNASSVEQALQILKEEQPKDYLLQHHNIRSNLERIFTKVPEPWVPPFHLSSFTHVPAEMQAWVDGYFGRGAAARPERYMSIIVEGDSRTGKTMWARSLGPHNYLMGHLDFNSRVYSNAAEYNVIDDISPNYLKLKHWKELIGAQRDWQSNCKYGKPVQIKGGIPSIVLCNPGEGASYKDFLGKEENRALHNWTIHNALFITFTAPLYQSTASPCQT |
| NCBI Accession | YP_001876453.1 |
|---|---|
| Location | 2074-2331 |
| Gene Name | AC4 |
| Protein Name | AC4 protein |
| Coding Region | ATGGGCAGCCTCATCTCCATGTGCTTATCCAGTTCGAAGGAAAATTCGTCCGCACGAATAACAGATTATTCGACCTGGTCTCCCCATCCCGGTCAGCACATTTCCATCCGAACATTCAGGGAGCTAAATCGAGCTCCGACGTCAAGTCCTATCTCGACAAGGATGGAGTCACAGTCGAATGGGGAGAATTCCAGGTCGACGGTCGAAGTGCTAGAGGAGGCTGCCAGAGTGCTAACGACTCATATGCCAAGGCATTGA |
| Protein Sequence | MGSLISMCLSSSKENSSARITDYSTWSPHPGQHISIRTFRELNRAPTSSPISTRMESQSNGENSRSTVEVLEEAARVLTTHMPRH |
| NCBI Accession | YP_001876454.1 |
|---|---|
| Location | 361-1128 |
| Gene Name | NSP |
| Protein Name | nuclear shuttle protein |
| Coding Region | ATGTCTTTTGTAAAATACAGGTATGGTGCATCATATACGACAAGACGTTTTTTACGTCAACAGGCTACTAAGAAAAGGAGTGCTGTTAAACGCAATGATTTTAAGCGTGGTTACAGACAAGTGAGCAAGTCTAATGAAGAGGCAAAGATGATTAGCCAATCATTGCATGAAAATCAGCTTGGTCCTGATTTTGTTATGACTCACAATAACGCTCTATCTACGTTCATTAATTTTCCATGTTTGGGTAAGACTTTACCCAACCGAAACAGGTCATATATTAAGTTGAAACGCTTGCGTTTCAAAGGTACGGTGAAGATAGAGCGTGTTCATGTGGATGTTAATATGGATGGTTTATCCCCTAAGATTGAAGGCGTATTTTCGCTTGTTATCGTCATCGACCGGAAACCACATCTTAACCCGAATGGCTGTCTACACTCATTTGATGAACTATTTGGAGCAAGGATACACAGCCATGGAAATCTTGCCATTACGTCGTCATTGAAAGATCGTTACTACATTCGTCATGTGTTCAAGAGTGTTATTTCTGTGATCAAAGACACTACAATGATCGACGTTGAGGGTTCCACTTTGTTGTCTAATAGGCGTTCGAACATGTGGTCGAGTTTTAAGGATCATGATCATGACTCTTGTAATGGTGTTTATGACAATATTAGCAAGAACGCCATATTAGTTTATTATTGTTGGATGTCCGACACCAACTCCAAGGCTTCCACTTTTGTATCGTTTGACCTTGACTATGTTGGATGA |
| Protein Sequence | MSFVKYRYGASYTTRRFLRQQATKKRSAVKRNDFKRGYRQVSKSNEEAKMISQSLHENQLGPDFVMTHNNALSTFINFPCLGKTLPNRNRSYIKLKRLRFKGTVKIERVHVDVNMDGLSPKIEGVFSLVIVIDRKPHLNPNGCLHSFDELFGARIHSHGNLAITSSLKDRYYIRHVFKSVISVIKDTTMIDVEGSTLLSNRRSNMWSSFKDHDHDSCNGVYDNISKNAILVYYCWMSDTNSKASTFVSFDLDYVG |
| NCBI Accession | YP_001876455.1 |
|---|---|
| Location | 1196-2077 |
| Gene Name | MP |
| Protein Name | movement protein |
| Coding Region | ATGGATTCTCAGTTAGCTTGTCCACCGAGTGCCTTTAATTATATTGAGTCTCATAGGGATGAATATCAGCTGTCTCATGATCTAACTGAGATAGTCCTTCAATTTCCTTCCACCGCTTCACAATTAACAGCTAGACTCAGTCGTAGTTGTATGAAAATCGACCATTGCGTCATAGAATATAGGCAACAGGTGCCTATCAACGCCACTGGAGCAGTCATAGTGGAGATTCATGACAAGAGAATGACGGACAACGAGTCCTTACAAGCTTCATGGACTTTTCCCATCCGGTGCAACATCGATCTCCACTATTTTTCATCGTCTTTCTTCTCGTTAAAAGACCCAATACCATGGAAACTCTATTACAGAGTTTCCGATACGAATGTCCATCAGAGAACACATTTCGCCAAATTCAAGGGAAAACTGAAATTATCAACGGCCAAACATTCAGTTGATATCCCATTCAGGGCTCCAACAGTCAAGATCCTCTCAAAACAGTTTTCCAATAGAGATGTCGATTTTTCTCATGTGGACTATGGGCGGTGGGAAAGGAAATTGATCAGGTCCGCATCGACTAACCAGTTTGGACTACATGGCCCAATTGAATTAAAACCAGGAGAGTCTTGGGCCACAAAAAGTTCAATAGGAGTGTCTCATACCGATGCGGACTCTGAATTAGAAAACGCAATACACCCATACAGACAACTAAACAGGCTCGGAACAAGCGTCCTAGACCCGGGAGATTCAGCATCAATTGTAGGAGCACAGAGGGCCCAATCAAACATAACAATGTCAATGACCCAATTAAGCGAGATTGTTAAGGCGACAGTTCAAGAATGTATTAACAGTAATTGTATCCCTTCTCAGCCAAAGTCGTTGAAATAA |
| Protein Sequence | MDSQLACPPSAFNYIESHRDEYQLSHDLTEIVLQFPSTASQLTARLSRSCMKIDHCVIEYRQQVPINATGAVIVEIHDKRMTDNESLQASWTFPIRCNIDLHYFSSSFFSLKDPIPWKLYYRVSDTNVHQRTHFAKFKGKLKLSTAKHSVDIPFRAPTVKILSKQFSNRDVDFSHVDYGRWERKLIRSASTNQFGLHGPIELKPGESWATKSSIGVSHTDADSELENAIHPYRQLNRLGTSVLDPGDSASIVGAQRAQSNITMSMTQLSEIVKATVQECINSNCIPSQPKSLK |
References More References in PubMed
| 1 |
Macroptilium yellow mosaic virus, a New Begomovirus Infecting Macroptilium lathyroides in Cuba. Ramos PL, et al. Plant Dis. 2002 Sep;86(9):1049. doi: 10.1094/PDIS.2002.86.9.1049B. PMID: 30818538 |
|---|---|
| 2 |
Bracero V, et al. Plant Dis. 2003 Sep;87(9):1022-1025. doi: 10.1094/PDIS.2003.87.9.1022. PMID: 30812812 |
| 3 |
Idris AM, et al. Plant Dis. 1999 Nov;83(11):1071. doi: 10.1094/PDIS.1999.83.11.1071C. PMID: 30841284 |
| 4 |
Collins A, et al. Virus Res. 2010 Jun;150(1-2):148-52. doi: 10.1016/j.virusres.2010.03.008. Epub 2010 Mar 25. PMID: 20347895 |
| 5 |
First Report of Bean common mosaic virus in Cudrania tricuspidata in Korea. Seo JK, et al. Plant Dis. 2015 Feb;99(2):292. doi: 10.1094/PDIS-07-14-0678-PDN. PMID: 30699581 |
| 6 |
Batista JG, et al. Arch Virol. 2022 Jul;167(7):1597-1602. doi: 10.1007/s00705-022-05410-0. Epub 2022 May 14. PMID: 35562613 |
| 7 |
Idris AM, et al. Plant Dis. 2002 May;86(5):558. doi: 10.1094/PDIS.2002.86.5.558C. PMID: 30818684 |
| 8 |
Two Newly Described Begomoviruses of Macroptilium lathyroides and Common Bean. Idris AM, et al. Phytopathology. 2003 Jul;93(7):774-83. doi: 10.1094/PHYTO.2003.93.7.774. PMID: 18943157 |
| 9 |
Nanopore sequencing of a novel bipartite New World begomovirus infecting cowpea. Naito FYB, et al. Arch Virol. 2019 Jul;164(7):1907-1910. doi: 10.1007/s00705-019-04254-5. Epub 2019 Apr 10. PMID: 30972591 |
| 10 |
Roye ME, et al. Plant Dis. 1997 Nov;81(11):1251-1258. doi: 10.1094/PDIS.1997.81.11.1251. PMID: 30861729 |