Jatropha mosaic India virus
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_002822725.1 |
| Isolate | India: U.P., Lucknow |
| Release date | 2018/8/25 |
| Submitter | Snehi,S.K., Raj,S.K., Prasad,V. |
| Host | |
| Vector | |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
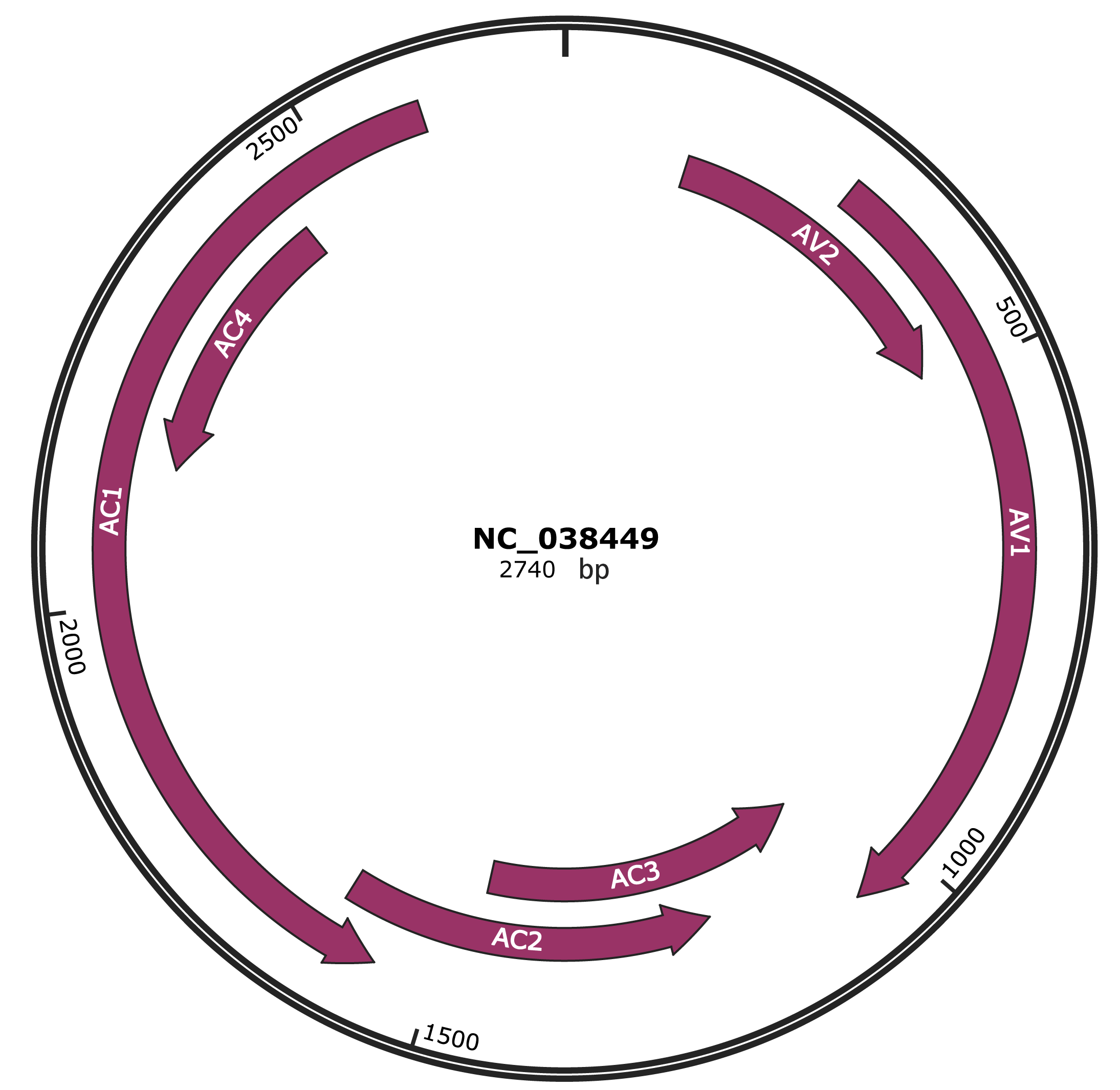
JBrowse
Genome
NC_038449
Gene Information
| NCBI Accession | YP_009506422.1 |
|---|---|
| Location | 135-491 |
| Gene Name | AV2 |
| Protein Name | pre-coat protein |
| Coding Region | ATGTGGGATCCACTACTTAACGAGTTCCCAGACTCCGTTCACGGTTTCCGTTGTATGCTAGCGCTGAAGTACCTTCAGCTCGTCGAAAGTACTTATTCCCCTGATACGCTCGGTTACGATCTAATCCGGGACCTGATTTCTGTTATCAGGGCCAGAAGCTATGTCGAAGCGACCAGGAGATATCATCATTTCAACTCCCGCCTCGAAGGTACGTCGCCGTCTGATCTTCGACAGCTTATACAGCAGCCGTGCTTCTGTCTTCACTGTCCGCGTCACCAAAAGACAAGCCTGGACAAACAGGCCCATGAATCGGAAGCCCAGATGGTACAGGATGTTCAGAAGCCCAGATGTTCCTAG |
| Protein Sequence | MWDPLLNEFPDSVHGFRCMLALKYLQLVESTYSPDTLGYDLIRDLISVIRARSYVEATRRYHHFNSRLEGTSPSDLRQLIQQPCFCLHCPRHQKTSLDKQAHESEAQMVQDVQKPRCS |
| NCBI Accession | YP_009506423.1 |
|---|---|
| Location | 295-1065 |
| Gene Name | AV1 |
| Protein Name | coat protein |
| Coding Region | ATGTCGAAGCGACCAGGAGATATCATCATTTCAACTCCCGCCTCGAAGGTACGTCGCCGTCTGATCTTCGACAGCTTATACAGCAGCCGTGCTTCTGTCTTCACTGTCCGCGTCACCAAAAGACAAGCCTGGACAAACAGGCCCATGAATCGGAAGCCCAGATGGTACAGGATGTTCAGAAGCCCAGATGTTCCTAGGGGATGTGAAGGCCCATGTAAGGTCCAGTCGTTTGAGTCCAGACACGATGTGGTCCATATAGGTAAGGTAATGTGCATCTCTGATGTCACTCGTGGTGTTGGGCTAACTCATCGTGTGGGTAAGAGGTTTTGCGTTAAGTCCGTTTACATCCTGGGGAAGGTATGGATGGATGAAAATATCAAGACCAAGAATCACACTAATAGCGTGATGTTCTTCCTCGTTAGGGATCGTAGGCCTGTTGATAAGCCCCAGGATTTCGGTGAGGTGTTTAACATGTTTGATAATGAACCTAGTACAGCTACTGTGAAGAACATGCATCGTGATCGCTATCAAGTCCTCAGGAAGTGGAGTGCCACTGTCACTGGTGGTCAGTATGCTAGCAAGGAGCAGGCTTTAGTTAGACGTTTTTTTAGAGTTAATAATTATGTTGTGTATAATCAGCAAGAGGCTGGCAAGTATGAAAATCATACTGAGAATGCATTGATGCTGTACATGGCGTGTACTCACGCCTCTAATCCTGTATACGCTACGCTGAAGATTAGAATCTACTTCTACGATTCGGTCAGCAATTAA |
| Protein Sequence | MSKRPGDIIISTPASKVRRRLIFDSLYSSRASVFTVRVTKRQAWTNRPMNRKPRWYRMFRSPDVPRGCEGPCKVQSFESRHDVVHIGKVMCISDVTRGVGLTHRVGKRFCVKSVYILGKVWMDENIKTKNHTNSVMFFLVRDRRPVDKPQDFGEVFNMFDNEPSTATVKNMHRDRYQVLRKWSATVTGGQYASKEQALVRRFFRVNNYVVYNQQEAGKYENHTENALMLYMACTHASNPVYATLKIRIYFYDSVSN |
| NCBI Accession | YP_009506424.1 |
|---|---|
| Location | 1062-1466 |
| Gene Name | AC3 |
| Protein Name | replication enhancer |
| Coding Region | ATGGATTCACGCACAGGGGAACTCATCACTGCAGCTCAGGCGATGAATGGCGTCTATACCTGGGAGGTTCCAAATCCCCTCTATTTCAAGATCATAGACCACAGCAGCCGGCCGTTCGGAACGACGATGGACATAATAACCGTCCAGATACGGTTCAACCACAACCTGAGGAAAGCGTTGGGACTACATCAATGCTGGATGGATTTCAAAATCTGGACGACCTTACGGCCTCAGACCTTGCGTTTCTTGAGGGTATTTAAGACCCACGTACTCAAATACCTGGACATGTTAGGGGTCATATGTATAAATACGGTTTTAACCGCCGTAGAACATGTATTGTACAGTGTACTCACAGGGACTGACAGTATTGAGCAGTCAAATTTAATAAAATTTAAAATTTATTAA |
| Protein Sequence | MDSRTGELITAAQAMNGVYTWEVPNPLYFKIIDHSSRPFGTTMDIITVQIRFNHNLRKALGLHQCWMDFKIWTTLRPQTLRFLRVFKTHVLKYLDMLGVICINTVLTAVEHVLYSVLTGTDSIEQSNLIKFKIY |
| NCBI Accession | YP_009506425.1 |
|---|---|
| Location | 1207-1614 |
| Gene Name | AC2 |
| Protein Name | transcription activator protein |
| Coding Region | ATGCGACCTTCATCTCCCTCGACGGGCCACTCTACTCCAGTGCCGATCAAGGTTAAACACAGGGCAGCTAAGCGTAGGGCCATCCGGCGACGGAGAGTAGACCTCAACTGCGGGTGCTCGTACTACGTGCACATCAACTGCCACAACCATGGATTCACGCACAGGGGAACTCATCACTGCAGCTCAGGCGATGAATGGCGTCTATACCTGGGAGGTTCCAAATCCCCTCTATTTCAAGATCATAGACCACAGCAGCCGGCCGTTCGGAACGACGATGGACATAATAACCGTCCAGATACGGTTCAACCACAACCTGAGGAAAGCGTTGGGACTACATCAATGCTGGATGGATTTCAAAATCTGGACGACCTTACGGCCTCAGACCTTGCGTTTCTTGAGGGTATTTAA |
| Protein Sequence | MRPSSPSTGHSTPVPIKVKHRAAKRRAIRRRRVDLNCGCSYYVHINCHNHGFTHRGTHHCSSGDEWRLYLGGSKSPLFQDHRPQQPAVRNDDGHNNRPDTVQPQPEESVGTTSMLDGFQNLDDLTASDLAFLEGI |
| NCBI Accession | YP_009506426.1 |
|---|---|
| Location | 1559-2602 |
| Gene Name | AC1 |
| Protein Name | replication-associated protein |
| Coding Region | ATGTCACCTCCTAAGCGTTTTATTATAAACGCCAAGAATTATTTCCTCACTTATCCCAGATGCTCTCTCTCCAAAGAAGAGACGCTTTCCCAAATTAAAAACCTAGAAACACCTACAAACCCTAAATTCATTAAAATCTGCAGAGAACTGCATGAGAATGGGGAACCTCATCTGCATGTGCTCGTCCAGTACGAGGGCAAGTTCAAATGCCAGAATAAGAGATTCTTCGACCTGGTATCCCCAACCAGGTCAGCACATTTCCATCCAAACATTCAGGGAGCTAAATCCAGCTCCGACGTCAAGTCCTACATCGACAAGGACGGAGATACTCTGGAATGGGGAGAATTCCAGGTCGACGGCAGGTCTGCACGGGGAGGCCAACAGTCGGCAAATGATGCATACGCCGCCGCAATTAACAGCGGCAGTAAGTCAGAGGCTCTTAAGATCCTTAGAGAGCTAGCTCCTAAAGACTACGTCCTGCAGTTTCATAATTTGAATGCCAATTTGGATAAAATTTTTACAACACCTGAAACTATCTATGAGAGTCCTTTTGGTCTGTCGACATTTGATCGAGTTCCAGAAGAACTCGATGTATGGTTCCATGAAAATGTCATGGATCCCGCTGCGCGGCCTTTGAGACCGAAGTCAATCGTAATCGAGGGCGATAGTCGGACAGGTAAGACTATGTGGGCTCGTGCATTGGGTCCACATAATTACCTATGTGGACATCTGGACTTGAGTCCTAGGGTCTACAGCAACGACGCGTGGTACAACGTCATTGATGACGTAGATCCCCACTACCTAAAGCACTTCAAGGAATTCATGGGGGCCCAGAAGGACTGGCAAAGCAACACCAAATACGGAAAGCCGGTTCAAATTAAAGGTGGGATACCCACTATCTTCCTGTGCAATCCTGGGCCCAATTCCAGCTATAAAGAGTTCCTTGACGAGCCGAAGAATACGGCATTGAAGGCTTGGGCAATAAGAAATGCGACCTTCATCTCCCTCGACGGGCCACTCTACTCCAGTGCCGATCAAGGTTAA |
| Protein Sequence | MSPPKRFIINAKNYFLTYPRCSLSKEETLSQIKNLETPTNPKFIKICRELHENGEPHLHVLVQYEGKFKCQNKRFFDLVSPTRSAHFHPNIQGAKSSSDVKSYIDKDGDTLEWGEFQVDGRSARGGQQSANDAYAAAINSGSKSEALKILRELAPKDYVLQFHNLNANLDKIFTTPETIYESPFGLSTFDRVPEELDVWFHENVMDPAARPLRPKSIVIEGDSRTGKTMWARALGPHNYLCGHLDLSPRVYSNDAWYNVIDDVDPHYLKHFKEFMGAQKDWQSNTKYGKPVQIKGGIPTIFLCNPGPNSSYKEFLDEPKNTALKAWAIRNATFISLDGPLYSSADQG |
| NCBI Accession | YP_009506427.1 |
|---|---|
| Location | 2143-2445 |
| Gene Name | AC4 |
| Protein Name | C4 protein |
| Coding Region | ATGGGGAACCTCATCTGCATGTGCTCGTCCAGTACGAGGGCAAGTTCAAATGCCAGAATAAGAGATTCTTCGACCTGGTATCCCCAACCAGGTCAGCACATTTCCATCCAAACATTCAGGGAGCTAAATCCAGCTCCGACGTCAAGTCCTACATCGACAAGGACGGAGATACTCTGGAATGGGGAGAATTCCAGGTCGACGGCAGGTCTGCACGGGGAGGCCAACAGTCGGCAAATGATGCATACGCCGCCGCAATTAACAGCGGCAGTAAGTCAGAGGCTCTTAAGATCCTTAGAGAGCTAG |
| Protein Sequence | MGNLICMCSSSTRASSNARIRDSSTWYPQPGQHISIQTFRELNPAPTSSPTSTRTEILWNGENSRSTAGLHGEANSRQMMHTPPQLTAAVSQRLLRSLES |
References More References in PubMed
| 1 |
First Report of Cucumber mosaic virus on Jatropha curcas in India. Raj SK, et al. Plant Dis. 2008 Jan;92(1):171. doi: 10.1094/PDIS-92-1-0171C. PMID: 30786384 |
|---|---|
| 2 |
Snehi SK, et al. Virus Genes. 2011 Aug;43(1):93-101. doi: 10.1007/s11262-011-0605-9. Epub 2011 Apr 9. PMID: 21479677 |
| 3 |
Gao S, et al. Arch Virol. 2010 Apr;155(4):607-12. doi: 10.1007/s00705-010-0625-0. Epub 2010 Mar 12. PMID: 20224893 |
| 4 |
Srivastava A, et al. Arch Virol. 2015 May;160(5):1359-62. doi: 10.1007/s00705-015-2375-5. Epub 2015 Feb 27. PMID: 25716923 |
| 5 |
Snehi SK, et al. Arch Virol. 2011 Dec;156(12):2303-7. doi: 10.1007/s00705-011-1118-5. Epub 2011 Oct 5. PMID: 21971870 |
| 6 |
Gireeshbai S, et al. Lett Appl Microbiol. 2022 Oct;75(4):1000-1009. doi: 10.1111/lam.13774. Epub 2022 Jul 2. PMID: 35723883 |
| 7 |
Srivastava A, et al. Virus Res. 2015 Apr 2;201:41-9. doi: 10.1016/j.virusres.2015.02.015. Epub 2015 Feb 23. PMID: 25720372 |
| 8 |
Wang G, et al. Virus Genes. 2014 Apr;48(2):402-5. doi: 10.1007/s11262-014-1034-3. Epub 2014 Jan 21. PMID: 24445901 |
| 9 |
Sidhu OP, et al. Planta. 2010 Jun;232(1):85-93. doi: 10.1007/s00425-010-1159-0. Epub 2010 Apr 7. PMID: 20372923 |
| 10 |
Kumar S, et al. Virusdisease. 2025 Jun;36(2):343-352. doi: 10.1007/s13337-025-00919-9. Epub 2025 May 8. PMID: 41018216 |