Jatropha leaf curl virus
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_000880535.1 |
| Isolate | India |
| Release date | 2015/2/22 |
| Submitter | Pal,A., Mukherjee,S. |
| Host | |
| Vector | |
| Download | Genome |GFF3 |PEP |CDS |
Genomic Organization
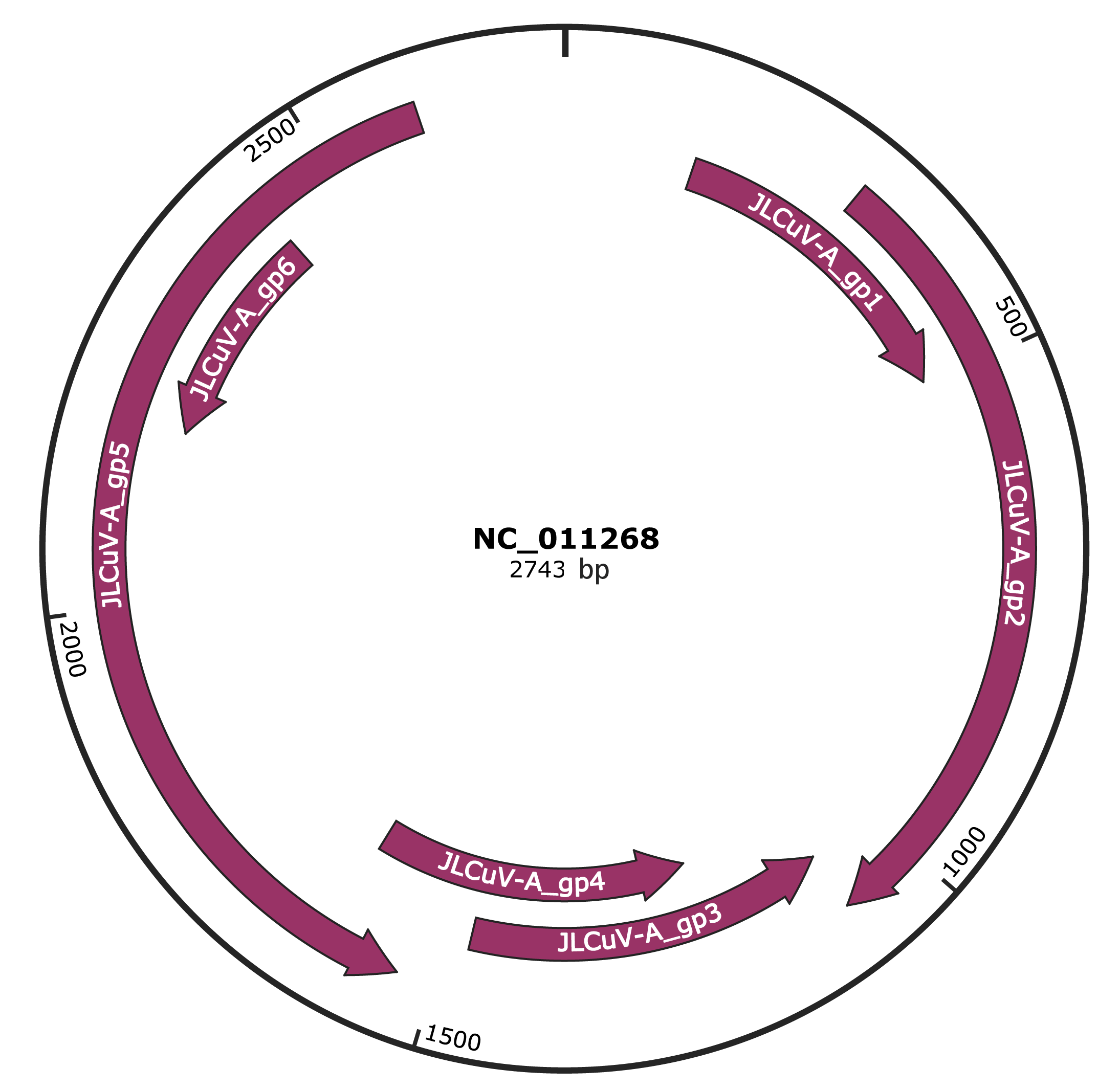
JBrowse
Genome
NC_011268
Gene Information
| NCBI Accession | YP_002224028.1 |
|---|---|
| Location | 143-496 |
| Protein Name | precoat protein |
| Coding Region | ATGTGGGATCCATTAACCAACGAGTTTCCCGAAACCCTGCACGGGTTTCGCTGTATGCTTGCAGTCAAATATCTACAAGCTCTACTCTCTTCGTACGCTCCTGATACGGTCGGATACGAGTACCTTCGTGATCTGATCAGCGTACTACGCTGTCGCGACTATGGCAAAGCGTCTATTCGATATGGTCGGATCGTCGCCGCTAAGAATAACACGCCGGAAACTGAACTTCGGGAGTCCCTCCCGTCTCATTGCCAGTGCCGTCACTGCCCCAGACATGGGATTCAAAAGGAGGGCGTGGACGTGGCGACCAGCAAACCGAAAACCGTGGATATACCGGAGAAGGAGAGCTCCTGA |
| Protein Sequence | MWDPLTNEFPETLHGFRCMLAVKYLQALLSSYAPDTVGYEYLRDLISVLRCRDYGKASIRYGRIVAAKNNTPETELRESLPSHCQCRHCPRHGIQKEGVDVATSKPKTVDIPEKESS |
| NCBI Accession | YP_002224029.1 |
|---|---|
| Location | 303-1079 |
| Protein Name | coat protein |
| Coding Region | ATGGCAAAGCGTCTATTCGATATGGTCGGATCGTCGCCGCTAAGAATAACACGCCGGAAACTGAACTTCGGGAGTCCCTCCCGTCTCATTGCCAGTGCCGTCACTGCCCCAGACATGGGATTCAAAAGGAGGGCGTGGACGTGGCGACCAGCAAACCGAAAACCGTGGATATACCGGAGAAGGAGAGCTCCTGATGTGCCGCGTGGATGCGAGGGACCATGCAAAGTACAGTCATTCGAAAAGAGACATGATGTCACGCACACGGGGGGCGTTCTATGTATATCTGACGTCACACGCGGCAATGGTATCACGCATAGAGTGGGTAAGAGATTCTGCGTGAAATCCGTCTATGTACTTGGCAAGATTTGGATGGACGAGAACATCAAGACGAAGAACCATACTAACAGCGTGATATTCTGGCTGGTCAGAGACAGGCGTCCATTTGGCAGTCCAATGGACCTGGGCCAAGTCTTTAACATGTATGATAACGAGCCCAGTACAGCCACTATCAAGAACGATTTGAGGGATAGATATCAAGTGTTACACAAATGGCACGCGTCGGTTACTGGTGGTGTCTATGCGTCTAAGGAACAAGCTATAATCAAGAAGTTCTACAGAGTCAATAATTATGTGGTCTACAATCACCAGGAAGCCGCGAAATACGAAAACCATACTGAGAATGCGTTATTATTGTACATGGCATGTACGCATGCATCAAATCCAGTGTATGCAACATTGAAAATACGCATATATTTCTACGACTCAGTATCAAATTAA |
| Protein Sequence | MAKRLFDMVGSSPLRITRRKLNFGSPSRLIASAVTAPDMGFKRRAWTWRPANRKPWIYRRRRAPDVPRGCEGPCKVQSFEKRHDVTHTGGVLCISDVTRGNGITHRVGKRFCVKSVYVLGKIWMDENIKTKNHTNSVIFWLVRDRRPFGSPMDLGQVFNMYDNEPSTATIKNDLRDRYQVLHKWHASVTGGVYASKEQAIIKKFYRVNNYVVYNHQEAAKYENHTENALLLYMACTHASNPVYATLKIRIYFYDSVSN |
| NCBI Accession | YP_002224030.1 |
|---|---|
| Location | 1076-1474 |
| Protein Name | AC3 |
| Coding Region | ATGGATTCACGCACAGGGGAACCCATCACTGCAGCTCGTTTCAACAATGGCGTCTTTATCTGGACGGTCCCAAATCCTCTGTACTTCCGCGTCATCAGTCACATGAGCAGACCCTTCAACCTGGACCAGGACATCATACACCTCCGAATACAATTCAATCACAATCTCAGGCAAGCACTACAGATACACAAGTGCTTCCTGAGCTTCAAAGTCTGGACTCGTTCACGGATTCAGACTGGGATGTTCTTACGAGTCTTTAAGACTCAAGTTATTAGATATTTAGATAGATTAGGTGTAATTTCTATTAATCTTGTAATTAAAGCTGTAGATCATGTATTGTATAATGTACTGCATCATACGATGCAGGTTGAACAGAGCAATGAAATAATGTTTAATTAA |
| Protein Sequence | MDSRTGEPITAARFNNGVFIWTVPNPLYFRVISHMSRPFNLDQDIIHLRIQFNHNLRQALQIHKCFLSFKVWTRSRIQTGMFLRVFKTQVIRYLDRLGVISINLVIKAVDHVLYNVLHHTMQVEQSNEIMFN |
| NCBI Accession | YP_002224031.1 |
|---|---|
| Location | 1215-1613 |
| Protein Name | AC2 |
| Coding Region | ATGTCGTGTTCGACAGCGACAGTCCCCCTCCAGCTCCTTTCAAGAGACGCCAGAGGCGACAGATCAGGGGGGGATAAGGCGGAGGAGGATCGACTTGAAGTGCGGATGTTCATATTTTCTCTCGCTAACTGCGCGAATCATGGATTCACGCACAGGGGAACCCATCACTGCAGCTCGTTTCAACAATGGCGTCTTTATCTGGACGGTCCCAAATCCTCTGTACTTCCGCGTCATCAGTCACATGAGCAGACCCTTCAACCTGGACCAGGACATCATACACCTCCGAATACAATTCAATCACAATCTCAGGCAAGCACTACAGATACACAAGTGCTTCCTGAGCTTCAAAGTCTGGACTCGTTCACGGATTCAGACTGGGATGTTCTTACGAGTCTTTAA |
| Protein Sequence | MSCSTATVPLQLLSRDARGDRSGGDKAEEDRLEVRMFIFSLANCANHGFTHRGTHHCSSFQQWRLYLDGPKSSVLPRHQSHEQTLQPGPGHHTPPNTIQSQSQASTTDTQVLPELQSLDSFTDSDWDVLTSL |
| NCBI Accession | YP_002224032.1 |
|---|---|
| Location | 1537-2601 |
| Protein Name | AC1 |
| Coding Region | ATGCCTCGTGTTAACGCATTTTCAGTTAGCGCTAAAAACATATTCCTCACTTATCCCAAATGCCCATTATCCAAGGAGACAGTCTTGGATCTCCTGCGAAACATATCCTGTCCTTCTGATAAATTATTTATTAGAGTCGCGCAGGAGAAACACGAGGATGGGTCACTGCATATCCATGCCCTTATCCAGTTCAAGGGTAAGGCCAGATTCAGAAATGCAAGACATTTCGATCTCATCCATCCTCATAGCTCCTCCCAATTCCATCCAAATATCCAGGGAGCTAAGTCCTCCTCTGATGTTAAGTCCTATATCGAGAAGGACGGAGATTTCATCGACTGGGGGTTCTTTCAGATCGATGGAAGATCTGCTAGAGGAGGTCAACAGACAGCTAATGATGCTGCAGCGGAGGCCTTAAACTCTGGGTCTGCAGAAGCTGCTCTAGCTATATCCGTGAAGATGCCCAGAGATTATATTTTTCAGTATCATAATTTAAAGTCCAATCTAGAGAGGGAGATATTTACACCTCTTAGAGTAGATTATGTTTCTCCTTATCCTCTTTGTTCGTTTGATCGAGTTCCAGAAGAACTCGAGATTTGGGCCTCAGAAAATATTGTCAGTCCCGCTGCGCGGCCTTTTAGACCCATAAGTATTGTTTTAGAGGGTGATAGTCGTATTGGGAAGACAATGTGGGCTCGGTCTCTCGGCCCACACAATTATTTATGCGGTCACTTGGATCTCAGTCCAAGAGTCTATAGTAATGACGTATGGTATAACGTCATTGATGATGTTGATCCGCATTATCTCAAGCACTTCAAGGAATTTATGGGGGCCCAGAGAGACTGGCAAAGCAACACAAAGTATGGAAAGCCCGTTCAAGTTAAAGGAGGCATTCCGACAATCTTCCTCTGCAATCCTGGGCCCCATTCGAGCTATAAAGAGTATTTAGACGAAGATAGGAACGCAGCATTAAAGAACTGGGCTGTTAAGAATGTCGTGTTCGACAGCGACAGTCCCCCTCCAGCTCCTTTCAAGAGACGCCAGAGGCGACAGATCAGGGGGGGATAA |
| Protein Sequence | MPRVNAFSVSAKNIFLTYPKCPLSKETVLDLLRNISCPSDKLFIRVAQEKHEDGSLHIHALIQFKGKARFRNARHFDLIHPHSSSQFHPNIQGAKSSSDVKSYIEKDGDFIDWGFFQIDGRSARGGQQTANDAAAEALNSGSAEAALAISVKMPRDYIFQYHNLKSNLEREIFTPLRVDYVSPYPLCSFDRVPEELEIWASENIVSPAARPFRPISIVLEGDSRIGKTMWARSLGPHNYLCGHLDLSPRVYSNDVWYNVIDDVDPHYLKHFKEFMGAQRDWQSNTKYGKPVQVKGGIPTIFLCNPGPHSSYKEYLDEDRNAALKNWAVKNVVFDSDSPPPAPFKRRQRRQIRGG |
| NCBI Accession | YP_002224033.1 |
|---|---|
| Location | 2187-2426 |
| Protein Name | AC4 |
| Coding Region | ATGCCCTTATCCAGTTCAAGGGTAAGGCCAGATTCAGAAATGCAAGACATTTCGATCTCATCCATCCTCATAGCTCCTCCCAATTCCATCCAAATATCCAGGGAGCTAAGTCCTCCTCTGATGTTAAGTCCTATATCGAGAAGGACGGAGATTTCATCGACTGGGGGTTCTTTCAGATCGATGGAAGATCTGCTAGAGGAGGTCAACAGACAGCTAATGATGCTGCAGCGGAGGCCTTAA |
| Protein Sequence | MPLSSSRVRPDSEMQDISISSILIAPPNSIQISRELSPPLMLSPISRRTEISSTGGSFRSMEDLLEEVNRQLMMLQRRP |
References More References in PubMed
| 1 |
Gireeshbai S, et al. Lett Appl Microbiol. 2022 Oct;75(4):1000-1009. doi: 10.1111/lam.13774. Epub 2022 Jul 2. PMID: 35723883 |
|---|---|
| 2 |
More P, et al. 3 Biotech. 2022 Oct;12(10):275. doi: 10.1007/s13205-022-03306-z. Epub 2022 Sep 12. PMID: 36110567 |
| 3 |
More P, et al. Sci Rep. 2021 Jan 13;11(1):890. doi: 10.1038/s41598-020-79134-z. PMID: 33441589 |
| 4 |
Chauhan S, et al. Gene. 2018 Jul 20;664:37-43. doi: 10.1016/j.gene.2018.04.062. Epub 2018 Apr 22. PMID: 29684487 |
| 5 |
Simmonds-Gordon RN, et al. Arch Virol. 2014 Oct;159(10):2815-8. doi: 10.1007/s00705-014-2112-5. Epub 2014 May 29. PMID: 24872185 |
| 6 |
Nawaz-ul-Rehman MS, et al. PLoS One. 2012;7(8):e40050. doi: 10.1371/journal.pone.0040050. Epub 2012 Aug 10. PMID: 22899988 |
| 7 |
Snehi SK, et al. Arch Virol. 2011 Dec;156(12):2303-7. doi: 10.1007/s00705-011-1118-5. Epub 2011 Oct 5. PMID: 21971870 |
| 8 |
Kumar S, et al. Virusdisease. 2025 Jun;36(2):343-352. doi: 10.1007/s13337-025-00919-9. Epub 2025 May 8. PMID: 41018216 |
| 9 |
Srivastava A, et al. Virus Res. 2015 Apr 2;201:41-9. doi: 10.1016/j.virusres.2015.02.015. Epub 2015 Feb 23. PMID: 25720372 |
| 10 |
Kashina BD, et al. Arch Virol. 2013 Feb;158(2):511-4. doi: 10.1007/s00705-012-1512-7. Epub 2012 Oct 17. PMID: 23074041 |