Hollyhock yellow vein virus
Basic Information
| Genus | Begomovirus |
|---|---|
| NCBI Assembly | GCF_013088155.1 |
| Isolate | India |
| Release date | 2021/6/1 |
| Host | |
| Download | Genome |GFF3 |PEP |CDS |
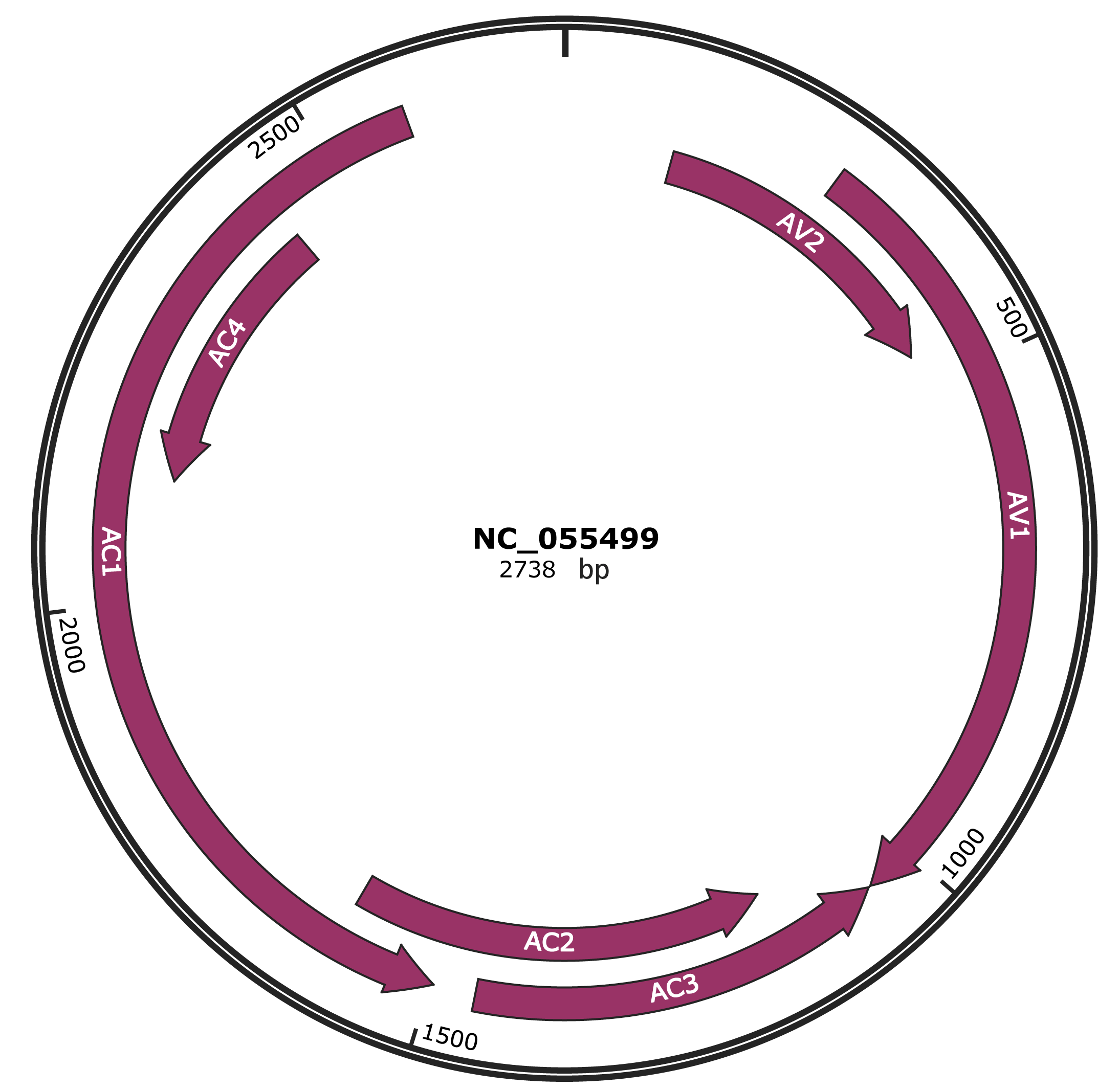
Genomic Organization
JBrowse
Genome
NC_055499
Gene Information
| NCBI Accession | YP_010086869.1 |
|---|---|
| Location | 118-465 |
| Gene Name | AV2 |
| Protein Name | movement protein |
| Coding Region | ATGTGGGATCCACTTTTAAACGAGTTCCCTGATACGGTTTACGGTTTTCGGTGTATGCTTTCTGTCAAATACTTGCAGCTATTGTCTGAAGGATACTCTCCGGATACGGTTGGGTACGAGTTAATTCGCGATCTAATTACTATTGTCCGATCCCGGAATTATGTCGAAGCGTCCTGCAGATATCGTAATTTCTACACCCGCGTCGAAAGTACGCCGGCGTCTGAACTTCGACAGCCCTTACGCGACCCGTGCTGGTGTCCCCACTGTCCGCGTCACAAAGTCAAGGCTATGGGCGAACAGGCCCATGAATCGGAAGCCCAGAATGTACAGAATGTACAGAAGCCCTGA |
| Protein Sequence | MWDPLLNEFPDTVYGFRCMLSVKYLQLLSEGYSPDTVGYELIRDLITIVRSRNYVEASCRYRNFYTRVESTPASELRQPLRDPCWCPHCPRHKVKAMGEQAHESEAQNVQNVQKP |
| NCBI Accession | YP_010086870.1 |
|---|---|
| Location | 278-1048 |
| Gene Name | AV1 |
| Protein Name | coat protein |
| Coding Region | ATGTCGAAGCGTCCTGCAGATATCGTAATTTCTACACCCGCGTCGAAAGTACGCCGGCGTCTGAACTTCGACAGCCCTTACGCGACCCGTGCTGGTGTCCCCACTGTCCGCGTCACAAAGTCAAGGCTATGGGCGAACAGGCCCATGAATCGGAAGCCCAGAATGTACAGAATGTACAGAAGCCCTGATGTTCCAAGGGGCTGTGAGGGTCCATGTAAGGTACAGTCATTTGAGTCCAGACACGATGTAGTCCATATAGGTAAAGTCATGTGTATTAGTGATGTTACTCGCGGTACTGGGCTGACCCATAGAGTTGGGAAACGGTTTTGTGTGAAGTCAGTCTATGTATTGGGTAAGATCTGGATGGATGAAAATATAAAGTCCAAGAATCATACTAACAATGTGATGTTCTTCCTCGTTCGAGATCGACGTCCCACCGGCACGCCTCAAGATTTTGGAGATGTCTTCAACATGTTTGACAACGAGCCTAGCACTGCTACTGTGAAGAATGTCCATCGGGATCGTTACCAGGTCTTGAGGAAATGGTATGCAACTGTCACTGGTGGACAGTATGGTGCAAAGGAACAGGCTTTAGTTAAGAAGTTTGTTAGGGTTAATAATTATGTTGTTTACAACCAGCAAGAGGCAGGGAAATACGAGAATCATTCTGAGAATGCCCTGATGTTGTATATGGCATGTACTCATGCCTCTAATCCTGTTTACGCGACTCTTAAGATTAGGATCTACTTTTATGACTCTGTAACGAATTGA |
| Protein Sequence | MSKRPADIVISTPASKVRRRLNFDSPYATRAGVPTVRVTKSRLWANRPMNRKPRMYRMYRSPDVPRGCEGPCKVQSFESRHDVVHIGKVMCISDVTRGTGLTHRVGKRFCVKSVYVLGKIWMDENIKSKNHTNNVMFFLVRDRRPTGTPQDFGDVFNMFDNEPSTATVKNVHRDRYQVLRKWYATVTGGQYGAKEQALVKKFVRVNNYVVYNQQEAGKYENHSENALMLYMACTHASNPVYATLKIRIYFYDSVTN |
| NCBI Accession | YP_010086871.1 |
|---|---|
| Location | 1051-1455 |
| Gene Name | AC3 |
| Protein Name | replication enhancer protein |
| Coding Region | ATGGATTCACGCACAGGGGAACCCATCACTGCAGCTCAAGCATGGAATGGCGTGTATATCTGGGACGTACCAAATCCCCTTTATTTCAAGATCCTAAGCCACTACGAGGCTCCATCAACATCGAGAATGACGATAATCACCATCAGGATCCAGTTCAATCACAACCTGAGGAAAGCACTGGGGATGCACAAGGGTTTTCTGACCTACCGAATCTGGACGACCTTACACCCTCCGATTGGTCTTTTCTTAAGAGTATTCCGAACCCAAGTCCTCAAGTATCTCAACAATCTAGGTGTAATTTCACTTAATTTTGTAATTAGGGCAATAAATCATGTATTGTGGAATGTAATTGCTCATACTGTAGAAGTGCAACAACTTTCAGAAATAAAATTCAAATTATATTAA |
| Protein Sequence | MDSRTGEPITAAQAWNGVYIWDVPNPLYFKILSHYEAPSTSRMTIITIRIQFNHNLRKALGMHKGFLTYRIWTTLHPPIGLFLRVFRTQVLKYLNNLGVISLNFVIRAINHVLWNVIAHTVEVQQLSEIKFKLY |
| NCBI Accession | YP_010086872.1 |
|---|---|
| Location | 1148-1600 |
| Gene Name | AC2 |
| Protein Name | transcription activator protein |
| Coding Region | ATGCACTCTTCTTCACCCTCGAGGAACCACTATACTCAAGTGCCGATCAAAGTCCAACACAGAGAGGCCAAGAAGAGGATCAGACGGAGGAGAGTGGATCTTGATTGCGGCTGTTCATATTACCTATCCATCAACTGCCACGATCATGGATTCACGCACAGGGGAACCCATCACTGCAGCTCAAGCATGGAATGGCGTGTATATCTGGGACGTACCAAATCCCCTTTATTTCAAGATCCTAAGCCACTACGAGGCTCCATCAACATCGAGAATGACGATAATCACCATCAGGATCCAGTTCAATCACAACCTGAGGAAAGCACTGGGGATGCACAAGGGTTTTCTGACCTACCGAATCTGGACGACCTTACACCCTCCGATTGGTCTTTTCTTAAGAGTATTCCGAACCCAAGTCCTCAAGTATCTCAACAATCTAGGTGTAATTTCACTTAA |
| Protein Sequence | MHSSSPSRNHYTQVPIKVQHREAKKRIRRRRVDLDCGCSYYLSINCHDHGFTHRGTHHCSSSMEWRVYLGRTKSPLFQDPKPLRGSINIENDDNHHQDPVQSQPEESTGDAQGFSDLPNLDDLTPSDWSFLKSIPNPSPQVSQQSRCNFT |
| NCBI Accession | YP_010086873.1 |
|---|---|
| Location | 1497-2585 |
| Gene Name | AC1 |
| Protein Name | replication associated protein |
| Coding Region | ATGCCTCCAAAGCGTTTTCTTATTAATTCCAAAAATTATTTCCTCACTTATCCGAAATGCTCTCTTACAAAAGAGGAAGCACTTTCCCAACTTCAAAACCTACAAACCCCAACAAATAAAAAATTCATCAAAATCTGCAAAGAGCTTCACGAGAATGGGGAACCTCATCTTCACGTGCTCATCCAGTTCGAGGGGAAATACAAGTGCCAGAATCAGCGATTCTTCGACCTGGTGTCCCCAAGCAGGTCAATACATTTCCATCCAAACATTCAGGGAGCTAAATCCAGCTCCGACGTCAAGTCCTATATCGACAAGGACGGAGACACCCTCGAATGGGGAGAGTTTCAGATCGATGGAAGATCTGCAAGAGGAGGACAACAGACAGCAAATGATGCTTACGCCCAGGCACTTAACACCGGCAGTAAGCCAGAGGCTCTTAGAGTACTTAGGGAGTTAGCTCCCAAAGATTATGTTTTACAATTTCATAATTTAAATGCCAATTTAGATAGGATTTTTACACCTCCCTTGGAGGTTTATGTTTCTCCTTTTTTATCTTCTTCTTTCAATCAAGTTCCAGAAGAACTTGAGGAATGGGTATCGGAAAACGTCATGGACGCCGCTGCGCGGCCTCTGCGTCCTTTGAGTTTAGTGTTAGAGGGAGACAGTCGTACGGGCAAAACGATGTGGGCACGGTCATTAGGACCACATAATTATCTTTGTGGCCATTTAGATCTGAGCCCTAAAGTCTACAGCAACGACGCGTGGTACAACGTCATTGATGACGTCGATCCGCACTTCCTCAAACACTTTAAAGAGTTCATGGGGGCCCAAAGGGACTGGCAATCAAACACGAAATACGGGAAGCCAGTTCAAATTAAAGGTGGGATCCCAACAATCTTCCTCTGCAATCCTGGGCCAAACTCATCGTATAAAGAGTACCTGGACGAGGAGAAGAACCAAGCCCTAAAAAACTGGGCTATTAAAAATGCACTCTTCTTCACCCTCGAGGAACCACTATACTCAAGTGCCGATCAAAGTCCAACACAGAGAGGCCAAGAAGAGGATCAGACGGAGGAGAGTGGATCTTGA |
| Protein Sequence | MPPKRFLINSKNYFLTYPKCSLTKEEALSQLQNLQTPTNKKFIKICKELHENGEPHLHVLIQFEGKYKCQNQRFFDLVSPSRSIHFHPNIQGAKSSSDVKSYIDKDGDTLEWGEFQIDGRSARGGQQTANDAYAQALNTGSKPEALRVLRELAPKDYVLQFHNLNANLDRIFTPPLEVYVSPFLSSSFNQVPEELEEWVSENVMDAAARPLRPLSLVLEGDSRTGKTMWARSLGPHNYLCGHLDLSPKVYSNDAWYNVIDDVDPHFLKHFKEFMGAQRDWQSNTKYGKPVQIKGGIPTIFLCNPGPNSSYKEYLDEEKNQALKNWAIKNALFFTLEEPLYSSADQSPTQRGQEEDQTEESGS |
| NCBI Accession | YP_010086874.1 |
|---|---|
| Location | 2129-2431 |
| Gene Name | AC4 |
| Protein Name | regulatory protein |
| Coding Region | ATGGGGAACCTCATCTTCACGTGCTCATCCAGTTCGAGGGGAAATACAAGTGCCAGAATCAGCGATTCTTCGACCTGGTGTCCCCAAGCAGGTCAATACATTTCCATCCAAACATTCAGGGAGCTAAATCCAGCTCCGACGTCAAGTCCTATATCGACAAGGACGGAGACACCCTCGAATGGGGAGAGTTTCAGATCGATGGAAGATCTGCAAGAGGAGGACAACAGACAGCAAATGATGCTTACGCCCAGGCACTTAACACCGGCAGTAAGCCAGAGGCTCTTAGAGTACTTAGGGAGTTAG |
| Protein Sequence | MGNLIFTCSSSSRGNTSARISDSSTWCPQAGQYISIQTFRELNPAPTSSPISTRTETPSNGESFRSMEDLQEEDNRQQMMLTPRHLTPAVSQRLLEYLGS |
References More References in PubMed
| 1 |
Khan A, et al. Arch Virol. 2021 Sep;166(9):2607-2610. doi: 10.1007/s00705-021-05134-7. Epub 2021 Jun 11. PMID: 34115211 |
|---|---|
| 2 |
First Report of Malva vein clearing virus Naturally Occurring in Hollyhock in Germany. Menzel W, et al. Plant Dis. 2010 Feb;94(2):276. doi: 10.1094/PDIS-94-2-0276B. PMID: 30754276 |
| 3 |
Srivastava A, et al. Arch Virol. 2014 Oct;159(10):2711-5. doi: 10.1007/s00705-014-2108-1. Epub 2014 May 9. PMID: 24810100 |
| 4 |
Kumar M, et al. Arch Virol. 2020 Sep;165(9):2099-2103. doi: 10.1007/s00705-020-04696-2. Epub 2020 Jun 18. PMID: 32556597 |
| 5 |
Azeem H, et al. Mol Biol Rep. 2022 Jun;49(6):5635-5644. doi: 10.1007/s11033-022-07557-0. Epub 2022 May 22. PMID: 35598198 |
| 6 |
Singh AK, et al. Plant Dis. 2021 Sep;105(9):2595-2600. doi: 10.1094/PDIS-12-20-2655-RE. Epub 2021 Oct 28. PMID: 33393356 |
| 7 |
Corchorus yellow vein virus, a New World geminivirus from the Old World. Ha C, et al. J Gen Virol. 2006 Apr;87(Pt 4):997-1003. doi: 10.1099/vir.0.81631-0. PMID: 16528050 |
| 8 |
Meena PN, et al. J Virol Methods. 2019 Jan;263:81-87. doi: 10.1016/j.jviromet.2018.10.016. Epub 2018 Oct 22. PMID: 30359678 |
| 9 |
Gupta K, et al. J Virol Methods. 2022 Feb;300:114413. doi: 10.1016/j.jviromet.2021.114413. Epub 2021 Dec 10. PMID: 34902462 |
| 10 |
Malvastrum yellow vein Yunnan virus is amonopartite begomovirus. Jiang T, et al. Acta Virol. 2010;54(1):21-6. doi: 10.4149/av_2010_01_21. PMID: 20201610 |